I know this error that I faced before. I guess you obtained Master_stack1_deb via stacking Master1_slave1+Master1_slave2… blah blah. Am I right? When I did same thing, I got same error too. Could you show us inside the deb and deb_insar products? When you get stack such I said your stack_deb file names like 2016_09_27_2016_09_27 that means combining more time. Look em all. And if I am right, just follow @bayzidul’s steps.
I am obtaining stack product using backgeocoding to Master+Slave1+Slave2+Slave3…etc after Orbit and Split each of them (Master and Slaves). Then I am debursting Stack product and I obtain stack_deb. Finally I am applying IFG that contain TOPO phase removel inside in the latest release (SNAP 6.0 Preview 4) and I get stack_deb_ifg_dinsar.
I hope u see.
Fikret
Hi jozep,
How did you solve the problem?
Filippo
Hi Filippo,
be sure to install tcsh package for cygwin and use script modified by @alvegavaleri in post 205.
Dear Jozep and all,
First of all I would like to thank you for your feedback. I have this problem in StaMPS step 3 when I’m running a stack of 4 scenes (in 1 patches); can you please tell me your suggestion regarding this problem?
Warning: Not enough random phase pixels to set gamma threshold - using default threshold of 0.3
In ps_select at 177
In stamps at 162
Thank you.
Best Regards,
Filippo
Hi everybody,
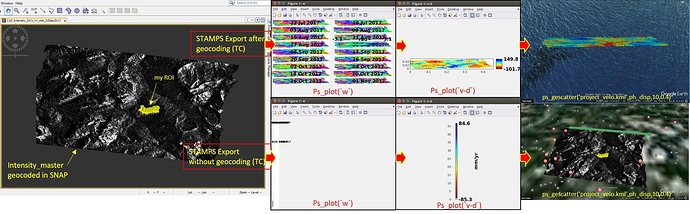
I managed to process 18 S-1 data using STAMPS but I encountered some problems and difficulties and I hope you can guide me. I used the mentioned guideline in this topic and exported STAMPS files using SNAP6.pr5 in two different ways : 1- with TC and 2- without TC (as you can see on the attached file).
When I did not use geocoded data in ‘‘STAMPS export’’ step, the wrap ifgs look wired but when I processed the data using the geocoded data (with TC) by STAMPS, at least I could have some results (of course I am not still sure regarding its correctness). After generating the kml file of ps, the ps pixels velocity located in the middle of the ocean (in ‘‘with TC’’ approach) and positioned at the top of my data as a line-like shape (in ‘‘without TC’’ approach)!!
It would be appreciated if anyone could help me.
Regards,
Mehdi
Dear all,
As you can see in my previous post I had the problem regarding the proper position of my PS pixels. I found the new version of SNAP by chance and in its Remove Topo tool has been added an ''oethorectified lat/long option (i.e. in the parameter section) as @mfitrzyk already mentioned. So, we do not need to geocode the needed files for STAMPS exporting anymore. My problem was solved by using the new version of SNAP (see below photo). I thought it worth sharing to save the time. Merry Christmas and happy New Year.
Hi all,
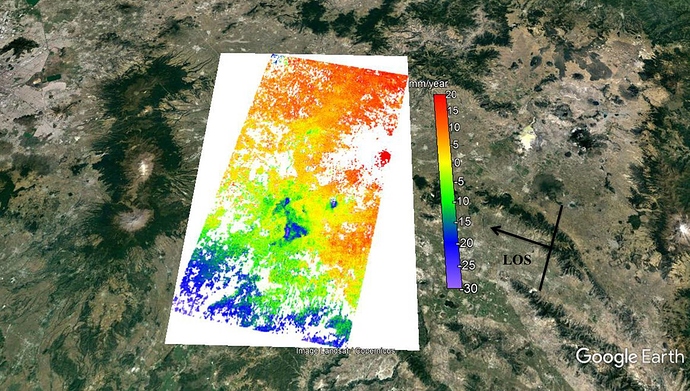
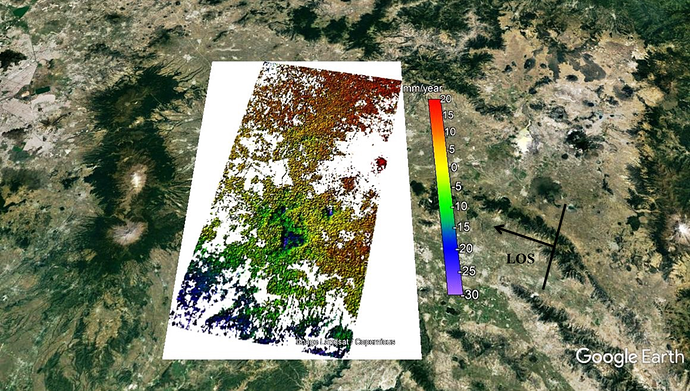
I processed 32 SLCs finally and the values of my results change between +20 and -30 mm/year (LOS velocity). I processed VV components and the negative values means subsidience after earthquake and but what does the mean of positive values ? I checked all unwrapped phases and the changes only occurs after earthquakes that bigger than 6.5 Mw. I showed normal and shaded relief verison of mean los velocity map. Thanks.
Fikret
Dear @Mehdi Im glad you have managed to process the data correctly.
Keep in ind that this version of the software is not an official release - just release candidate
The official release should be in the next couple of days!
Hi All
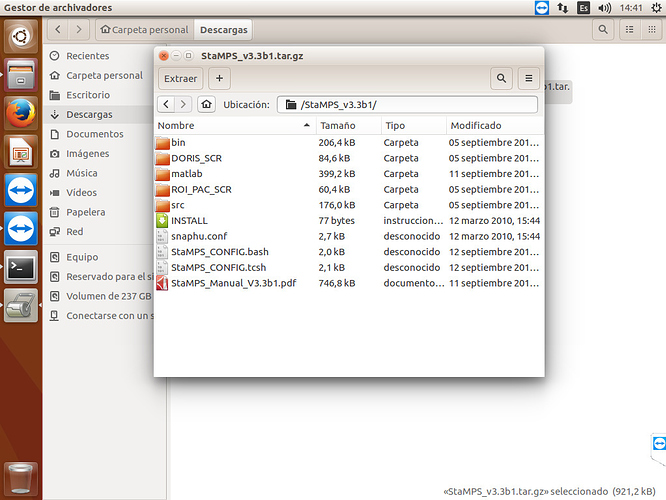
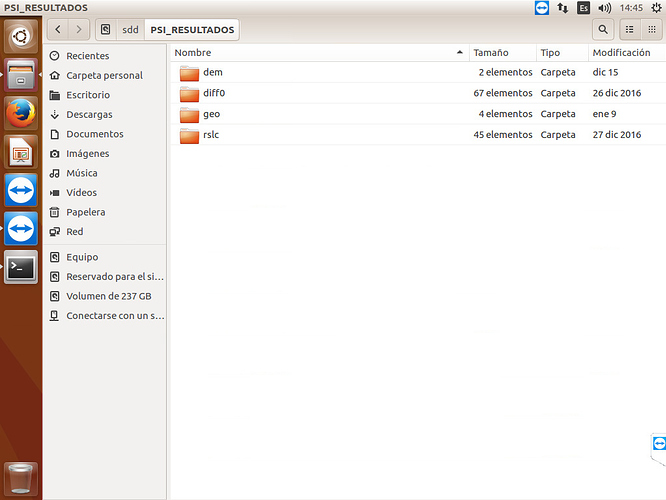
I don’t have the INSAR_master_date folder’s in my data of StaMPS.v3, I redirected all (geo, dem, dff0, rslc) in other folder and other directory… but when I run the command (mt_prep_gamma 20160208 0.4 2 2 50 200) this give me a error…
below print screen StaMPS.v3
my question is: Is necessary that all folder is in the same directory???
below print screen PSI_RESULTADOS
I don’t know if I understood correctly, but you should call mt_prep_gamma command from folder where your exported files are (from PSI_RESULTADOS). Before that you should add bin folder from StaMPS directory to path variables.
Hi Fikretjfm,
The velocity map you get is only a relative value, the zero values on the map don’t mean they don’t subsidence in reality, they are just the reference points. For example, if the subsidence velocity of the zero value points on the map is 20mm/yr in reality, then the real subsidence velocity of the points which value are -5mm/yr on the map is 15mm/yr, and the positive values on the map are calculated the same way.
All the best,
Fei
You are unable to run the command as the system doesn’t know where to look for the mt_prep_gamma script file.
Open a Terminal window.
To add the directory of the mt_prep_gamma script (in this case the StaMPS bin directory) to the path list type:
PATH=/yourStaMPSfolderdirectory/bin:$PATH.
You should now be able to execute the command.
To check the updated environment variable path list type:
echo $PATH
The newly added path should appear in the list.
To run the command and avoid any errors you can format it as follows:
mt_prep_gamma [yyyy/mm/dd - of master scene] [output_directory] 0.4 2 2 50 200
(given that you want to leave the default prep options)
Good luck
Thanks for your reply, before I make your suggested me, I have that to install the tcsh program!!! 
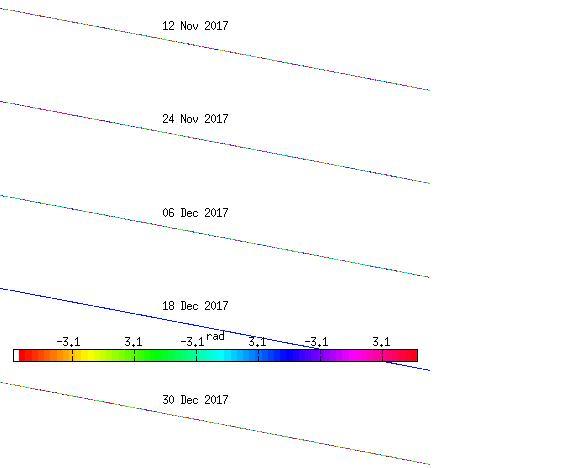
Hello everyone,
I followed the steps which Katherine provided and get weird wrapping result in StaMPS as below. Do you have any suggestion on this issue? Btw, unwrapping or plot any figure will have similar issue.
Many thanks.
#######################################################
Warning
The workflow described in this post (and the ones referring to it later on) is outdated and no longer recommended, because new versions of SNAP and StaMPS have been released. Please have a look at this updated list of instructions: StaMPS - Detailled instructions
#######################################################
Hi all,
Recently SNAP6.0 has been released, and by chance @Mehdi use the “oethorectified lat/long” options to solve his problem. And now I test this method and find that this new option can solve the shift problem and can avoid Terrain Correction (TC) step in PSI. @katherine @annamaria @bayzidul @ABraun
Here is the new Interferogram Formation window in SNAP6.0:
After finish this step, elevation and lat/lon band will be generated in result product.
Start from Deburst step, we get Stack_deb, then create a subset, get Subset_Stack_deb, form interfeorgrams as the first picture showed and get Subset_Stack_deb_ifg. And then use Subset_Stack_deb and Subset_Stack_deb_ifg to export to StaMPS.
And that’s all, all the things have been done. Add Elevation band, geocoded and topo_removal are doing together in interferogram formation step, TC are avoided, lat/lon file can export correctly without saving to GAMMA format.
Here are some results:
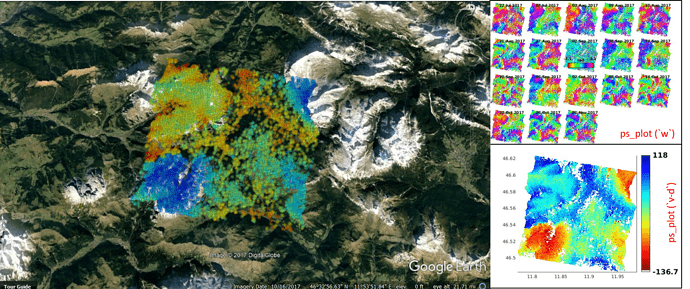
1.use SNAP6.preview4 and without TC:
2.use SNAP6.preview4 and TC:
3.use SNAP6.0 oethorectified lat/lon and without TC:
Note: I use different parameters when export to Google Earth, and only about 20% PSs are shown in the picture, so don’t be surprised that PSs location are not exactly the same.
In SNAP6.preview4 or preview5, without TC there are exist shift problem. And use SNAP6.0, shift problem has been solved and TC are no more needed.
Have Fun!
Fei
that’s just fantastic, thanks for testing!
One question about the stack: Did you create pairwise stacks of AB AC AD… and merge them with the back geocoding as originally suggested? Because this creates a quite redundant stack with A in it multiple times and I wondered if this is the best way to do it.
Dear ABraun,
I follow the step as katherine suggested here:
And my post can be seen as a suggestion for the orignal way.
Dear Fei,
Thank you so much for sharing your wonderful experience! Due to our cooperation the workflow between SNAP and StaMPS becomes more and more friendly! The next problem seems to be the possibility to create a stack of pairs with different masters (being resampled to one “supermaster”) and export such a stack to StaMPS to execute the StaMPS SB (Small Baselines Processing). StaMPS SB will be very useful in rural areas where StaMPS PS permits us to identify only few PS (especially using S-1 images)…
All the best,
Katherine