ok. It’s clear.
I do not know if these are PSI selected points (stable points).
Before to run stamps write in matlab’s terminal:
setparm (‘weedneighbours’, ‘n’)
(Open matlab, write setparm …, run stamps)
Do you have some change in stamps results?
Also, do you check in same way if dinsar interferograms stack are correct?
I’ve already set the weedneighbours parameters and changed the goldstein filter width. So yes these are the PSI selected points.
The Dinsar stack seems right, but it’s kind of hard to certify it. At least they do not appear abnormal.
Everything seems good… but I need furger investigation in order to be sure.
The only thing that surprise me a bit is the heading parameter which is set at 180.
I don’t know how help you. You could try to undestand if the results are consistent with ground truth, after we might talk again.
I think that is why, after TC, pixels coordinate are reprojected in latitude and longitude, not azimuth and range. I repete, this is my personal workflow, it could be affected by errors.
Let me know.
Hi MWampach,
I meet the same mistakes with you,could you please tell me how you sloved them?
Thanks a lot!
Hi juanesburgo,
I really don't konw the band you say which we should delete,could you please tell me the detial?
Thanks a lot!Hi Sharon,
I used the same two products(stack_deb_ifg and stack_deb_ifg_dinsar),the dem folder also didn’t appear,but when I
used the two products(stack_deb and stack_deb_ifg_dinsar),it would tell me a error that like “the product…already contains a band with the name’i_20151224_rslc’”.
I don’t konw have you solved ?Please tell me ,thank you!
#######################################################
Warning
The workflow described in this post (and the ones referring to it later on) is outdated and no longer recommended, because new versions of SNAP and StaMPS have been released. Please have a look at this updated list of instructions: StaMPS - Detailled instructions
#######################################################
Hi all,
@ABraun,@katherine,@cfatras
It seems that I have found a serious bug in mt_prep_gamma_snap, the file attached by Katherine.
If you open any *.rslc.par file in /rslc floder, you will see the parameter ‘image_format’ is ‘FCOMPLEX’, which is often 8 bytes in computer, meantime you can also get ‘range_samples’ and ‘azimuth_lines’ value, do multiplication between these three parameter and you will find it’s equal to the size of *.rslc exactly, which means the *.rslc only contains raw data.
Now we check the source code of mt_prep_gamma_snap in line 149 and line 230(or 232), all contains ‘s’ as an input parameter, which means all your data, including all *.rslc and *.diff, will be read as complex short format, which is often 4 bytes in computer, so, it means we have read the export data uncorrectly  .
.
Despite it will cause huge mistake, the solution is simple, just replace all ‘s’ as ‘f’, which will change the reading format to complex float, and then can read data correctly.
The reason why I find this problem is because the strange value in ‘selpsc.in’ file, from the third line, after the full path of *.rslc, a number appears.
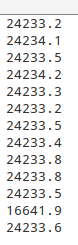
After I check the source code, I know it means mean amplitude in each *.rslc except zero amplitude pixel. So, over 20 thousand is totally unreasonable. After I modified the code and re-run mt_prep_gamma_snap, I think I get the right value.
Another clue is when you run mt_prep_gamma_snap, it will output a message in terminal, “number of lines per file = XXX”, it should tell you the length of *.rslc image, but you will find the output value equals twice of the real value, it because “numlines=size/width/sizeofelement/2”(in /src/selpsc_patch.c in line 264), and given ‘s’ as paramter will set sizeofelement equals 2.
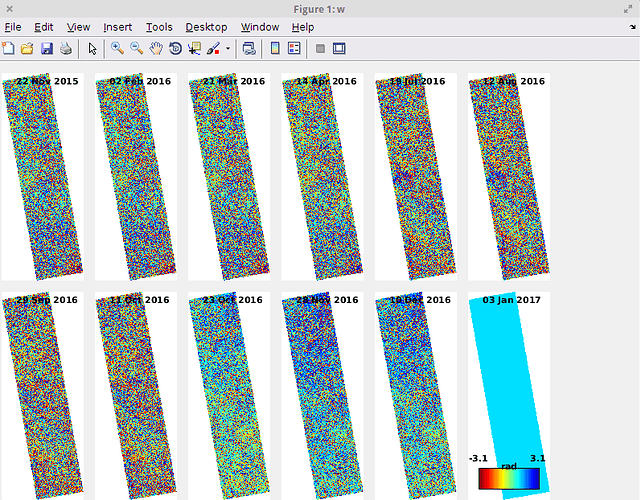
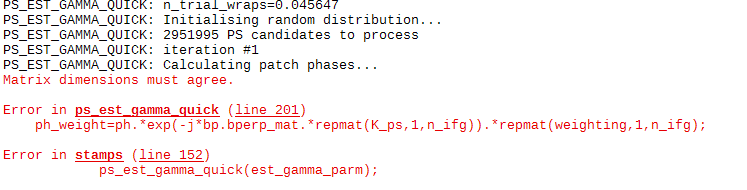
Actually I also have met another bugs in ps_load_initial_gamma.m, I have mentioned to ABraun once, but I don’t know if you guys have met the same problem. After run stamps(1, 1) in Matlab(which run ps_load_initial_gamma), use ps_plot(‘w’), I find that one of my ifg is empty, and I get an error in the following step stamps(2, 2).
Last ifg 03 Jan 2017 is empty
error message in step 2:
Later I find the reason is because the output data format of ps_load_initial_gamma is a little different from ps_load_initial(which is used for other sensor except gamma), mainly on the handling of master image, so I have modified it to solve this problem.
I don’t know if anyone has the same problems as I do, I have viewed the MAINSAR google group, also has no useful information about ps_load_initial_gamma.m.
At last, thanks for Katherine sharing her experience selflessly, I attach the modified code and change log here, hope this will help others and save their time.
Fei
Good luck!
Change_log.txt (1.2 KB)
mt_prep_gamma_snap(changed) (6.4 KB)
ps_load_initial_gamma(changed).m (5.8 KB)
great, thanks for sharing! I was also able to run until step 6 today but I am still experiencing. But I’ll definetly save your scripts and use them. Especially the first one… I often noticed the unnaturally high number of selected ps.
Wow, great work, thanks Fei!
I was digging into these strange values this morning, so thanks for sharing the solution with the community!
Dear Fei,
Thank you very much! Great job! I was also puzzled why I get so many PS! It is great when people help each other! Your changed scripts will save a lot of time to many people!
Thank you again!
All the best,
Katherine
Hi Adrian,
I’m trying from the beginning step. If I get the right folders successfully, I will that you know!
But I think you need to make interferogram before deburst, you may not get the error like this "The product … already contains a band with the name ‘i_201150408_rslc’.
And _Stack_ifg_deb_dinsar and _Stack_ifg_deb will be the products for stamps export.
Hi Sharon,
Thanks for your reply!You mean that your export folders also don’t include ‘dem’?
I will try making interferogram before deburst.And if I have new progress,I will reply.
Some weeks ago I found a little problem in K2q.m function: this script was written for other missions (Envisat, etc) and it isn’t been updated for Sentinel.
K2q is used in ps_output (ps_output permits to export ascii files of StaMPS elaborations) when the file is written dem_error.xy (in dem_error.xy file appears a column with all Nan values: it is caused by K2q “error”).
I changed ps_output for my purpose and i do not use K2q, so I can not attach a new version, but if someone modifies the script, he could attacch here.
Hi annamaria,
Thanks for your reply, you are right about the K2q.m function, I have also noticed that in many place of the source code, parameter or constant was written for other missions, I’m reading the source code now, maybe after that, I will update the source code and tell why we should change them. Annamaria, since you have changed the ps_outpt, could you please attach the modified code so that I can use as a reference?
But now I meet another problem, do anyone have any learning materials about PSI? Because I find sometimes it’s hard to understand the source code, the materials I found is either too simple or too complicated.
All the best,
Fei
Hi FeiLiu, thanks for your reply.
I changed code writing in italian and including other thinghs (for example a column for master image, that didn’t appeared). Now i haven’t time for to review the code, but i think that next week i’ll do. i’ll let you know.
I agree. Besides the StaMPS manual there are only few materials which assist self-teaching DInSAR with PSI.
What I found quite helpful was this review paper by Crosetto et al. (2016):
It’s not complete but it answered some of my questions.
Dear Fei,
This is the paper describing the main StaMPS algorithms. Alcedo.pdf (1.6 MB)
Good luck,
Katherine
Using Katherine instructions I were able to do the StaMPS export, but I´m having problems with StaMPS step 3, I get the following error:
PS_SELECT: Initial gamma threshold: 0.018 at D_A=0.06 to 0.219 at D_A=0.40
PS_SELECT: 41837 PS selected initially
Subscript indices must either be real positive integers or logicals.
Error in ps_select (line 277)
ph_bit=pm.ph_grid(i_min:i_max,j_min:j_max,:);
Error in stamps (line 162)
ps_select;
I found a thread in Mainsar forums here: https://groups.google.com/forum/#!topic/mainsar/jH9q6TtGXC8 I will try to increase the size of my subset (4710 x 305) but I don´t think that this is the case. Anyone is having the same issue?
Thanks!
Hi Sharon,
I have tried two days in different ways,but I still can’t export the correct folders that contain the’dem’ folders.Do you have some new progress?
I’m looking forward to your reply.