Thank you  @gabrielaquintana77
@gabrielaquintana77
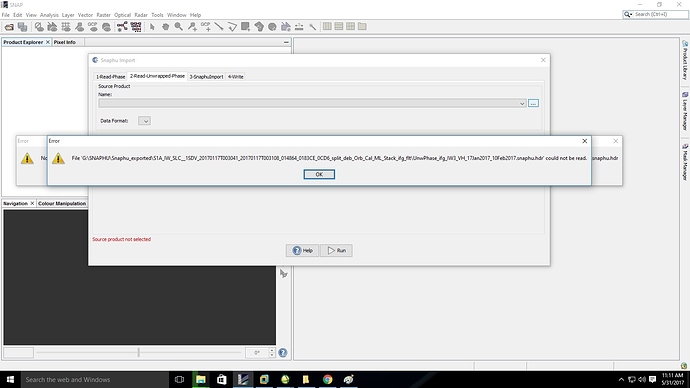
@lveci I have performed phase unwrapping using ubuntu through VMWARE. I am facing a problem while trying to import the unwrapped image.
Please help me resolve this issue.Importing the unwrapped interferogram should be performed with the import function in the interferometry module
thank you @ ABraun , I have tried importing using snaphu import, the same error still persists. kindly guide me through.
did you select the right reference file (1-Read-Phase) in the import module? This should actually copy the source image’s metadata into the imported unwrapped phase.
1-Read-Phase (this is the reference file, usually the one before the unwrapping)
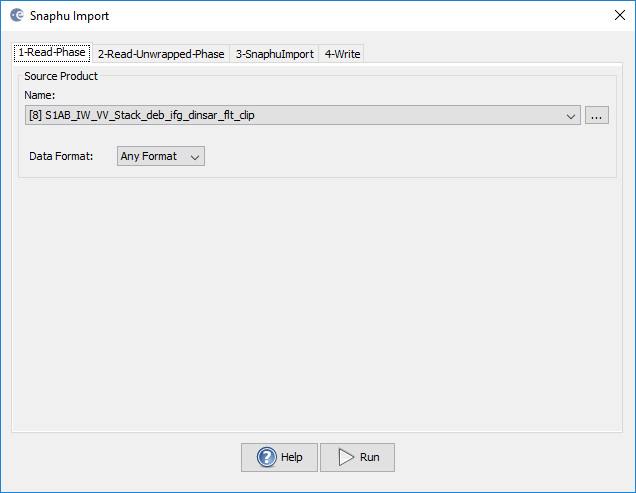
2-Read-Unwrapped-Phase (here you chose the hdr file of the unwrapped phase created by snaphu)
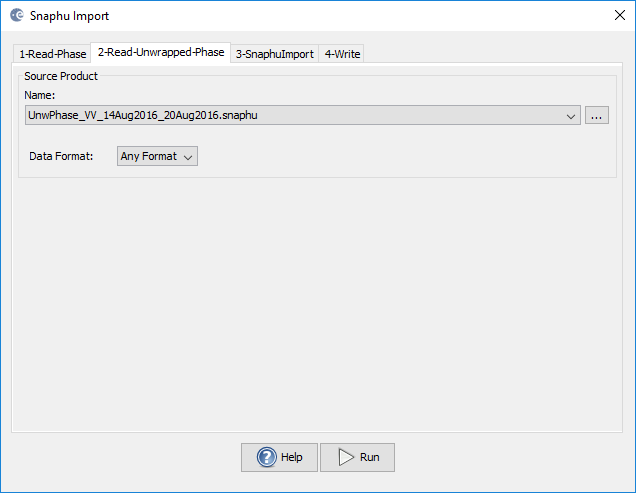
I have implemented the steps as suggested by you
I am still not able to figure where I am going wrong
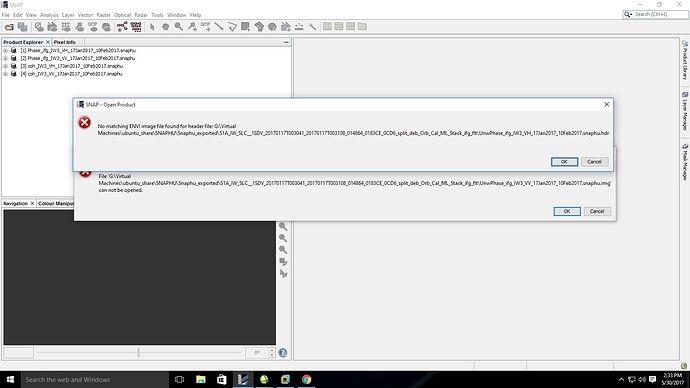
the second screenshot shows that your unwrapped phase was not generated. Only the metadata file (*.hrd) exists.
In this case snaphu did not run correctly, no unwrapped interferogram was created. Thus, there is nothing to import.
Just importing the header (*hdr) file without any image data won’t work.
Did you install and run snaphu successfully?
@ABraun , I have processed the image using ubuntu through VMWARE. this is the screenshot of the completed process
the message is displayed as ‘program snaphu done’ . so I am not sure if the image is completely unwrapped or if I should run it again.I would greatly appreciate your valuable suggestion.
The message indicates that everything went fine but in your screenshot the img file is not there.
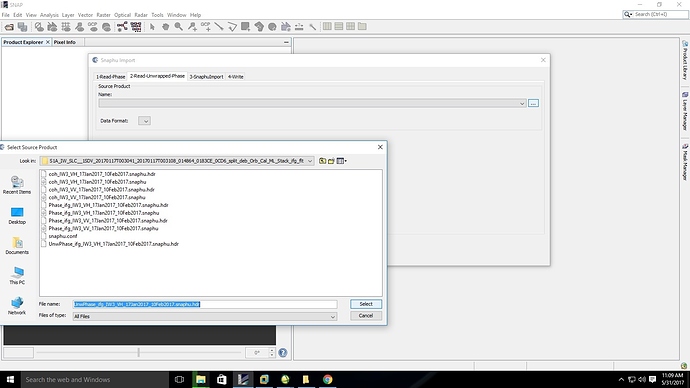
@ABraun, I have tried running snaphu again using MINT64 through VMWARE. while trying to import the image an error message is displayed as ‘unable to read the hdr file’ . Basically I found out that a header is created for ‘VH’ and an image file is created for ‘VV’,
could you please help me find out the error.have experienced something similar. I have no solution but deleting the VH bands before SNAPHU export. It is usually not of better quality and in the end you can only use one anyways.
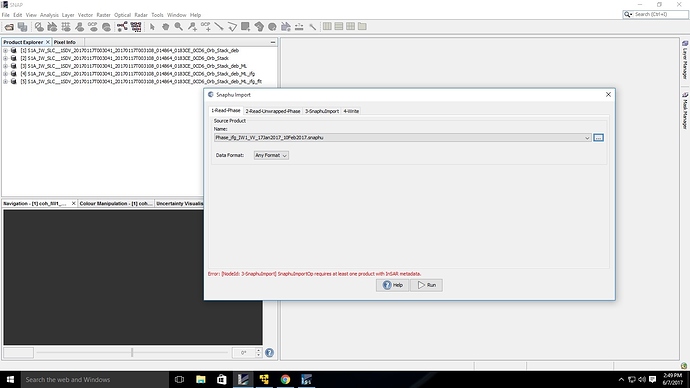
@ABraun I have implemented your suggestion of deleting VH band and ran snaphu for only VV band. when I amtrying to import in SNAP, I get an error saying ’
snaphuimportrequires atleast one product with InSAR metadata.Please help me solve this error.
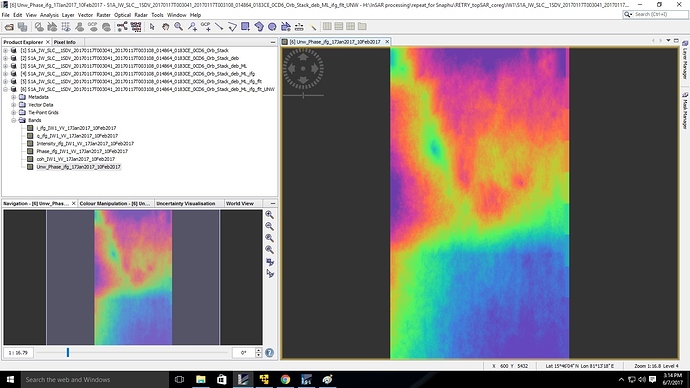
thank you, I have imported the file with metadata and succesfully generated imported unwrapped phase image.
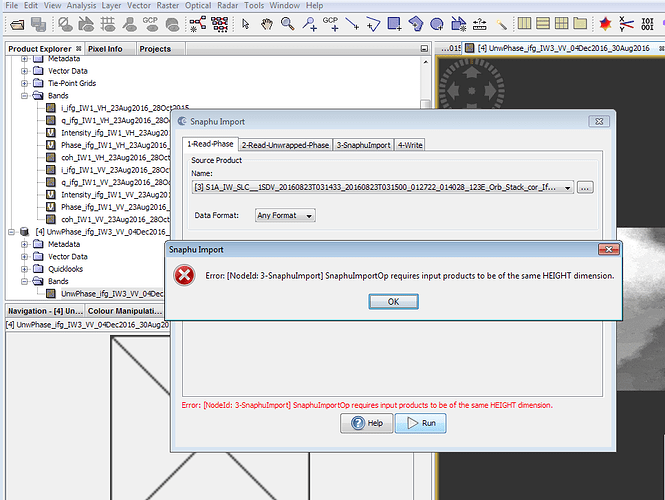
here is a screenshot of the same. I am not sure how good is the result because when I am trying the option “phase to elevation” , a java null pointer error shows up. could you please help me with this errorjust read the error message  The Product of 1-Read-Phase and 2-Read-Unwrapped-Phase must be of the same extent, that means they must have the same pixel numbers (lines and rows).
The Product of 1-Read-Phase and 2-Read-Unwrapped-Phase must be of the same extent, that means they must have the same pixel numbers (lines and rows).
Don’t open the unwrapped interferogram in SNAP before the import. Just select your stack before the unwrapping under tab 1 and select the unwrapped interferogram (*.hdr) in tab 2.
Hi lveci
i want to create a dem and follow all the steps mention in the forum. I use a single sub-swath but phase to elevation show java null pointer. Can you tell me the how can i solve the problem. I use Sentinel 1 vv data and middle sub-swath
Hi Fran,
I tried snaphu unwrapping with cygwin command line but i got an error. I am using snap on windows 7. I changed the current working directory but when i tried to run the config file, It says like
trainee2017228@MILABDHPE830015 ~
$ cd ‘/cygdrive/c/Users/trainee2017228/Desktop/New folder/snaphu/target_ifg_ML’
trainee2017228@MILABDHPE830015 /cygdrive/c/Users/trainee2017228/Desktop/New fold er/snaphu/target_ifg_ML
## $ -bash: snaphu: command not found
Please help me with the steps to complete the process.
Thank you