Hello,
I am also having some issues with Snaphu importing, I created an interferogram for an area on the west coast of Ireland using two SLC IW products. I exported the data from Snap and used Snaphu to perform the phase unwrapping. The screenshot below shows the wrapped phase (left) and the unwrapped phase (right), just wondering does the unwrapped phase image look correct as there does seem to be some areas where the image looks wrong.
Many thanks,
Darragh
looks fine to me. Of course areas with low coherence cannot be used and the nodata values are not supported by snaphu but in general, the result is correct.
hi all
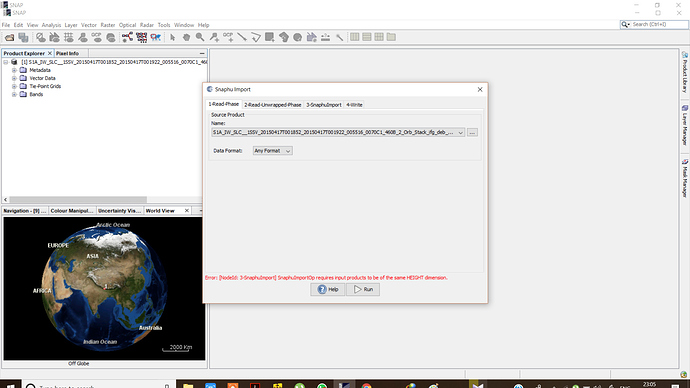
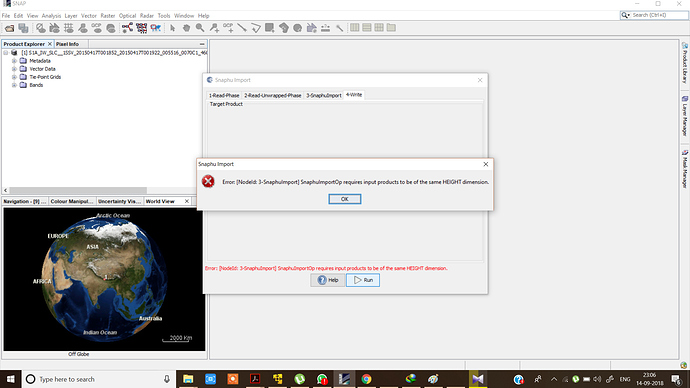
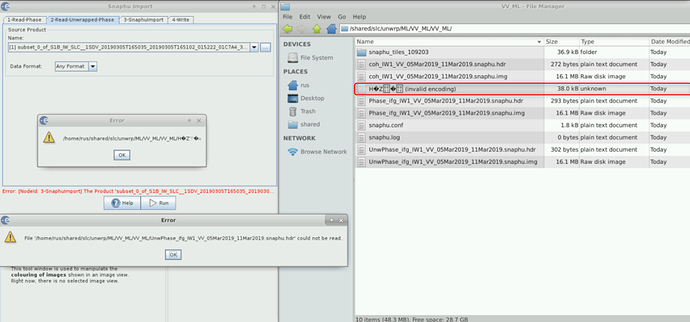
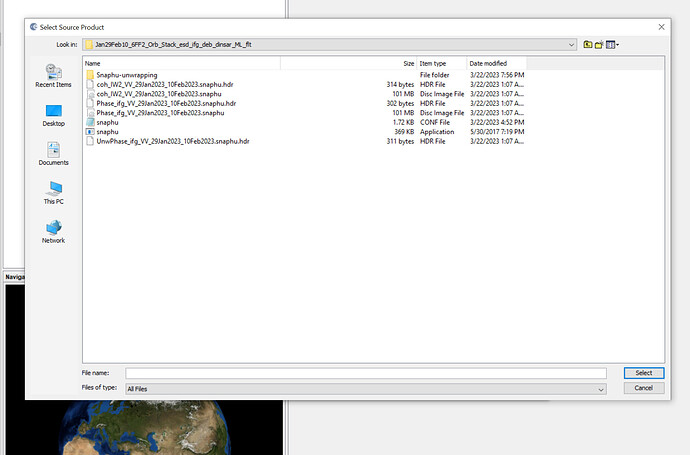
I want to generate DEM but when I am importing the Unwrapped phase with wrapped i am getting following problem each and every time.
I am importing it with deb_flt as a unwrapped file and .hdr as a unwrapped file
Hi @pratyusha I have been struggling with the same problem you had. I am unable to import the snaphu file despite the process was successfully completed.
Can you please write to me your exact step you took in order to get it working.
Thanks
hi, I have experienced the same problem as you had experienced … tell me how you solve it … coz I cant produce steps further becoz this issue … heip me to solve it … i send you the clips of the error message
Hi,
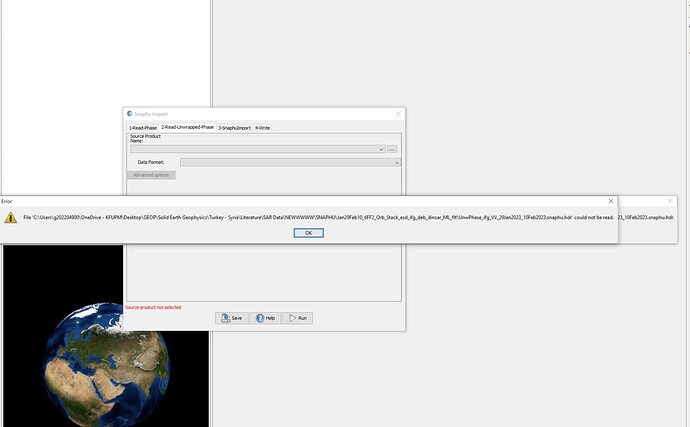
I am also having troubles with import of .hdr file.
Not only in snaphu import section but in general. This error shows up when I tried to open the file.
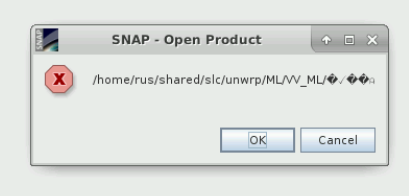
Is there any other problem in the data or settings?
Cheers
K.
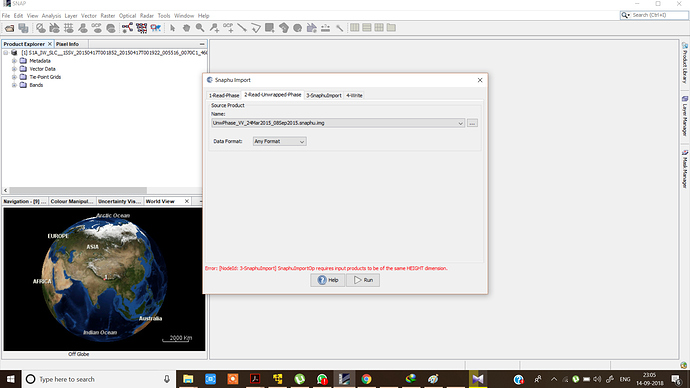
In the snaphu import choose your unwrapped_phase.img instead of the header file. This shall solve your issue
In the end, it was solved out.
There was some file that caused the problem. After I deleted it, all went well.
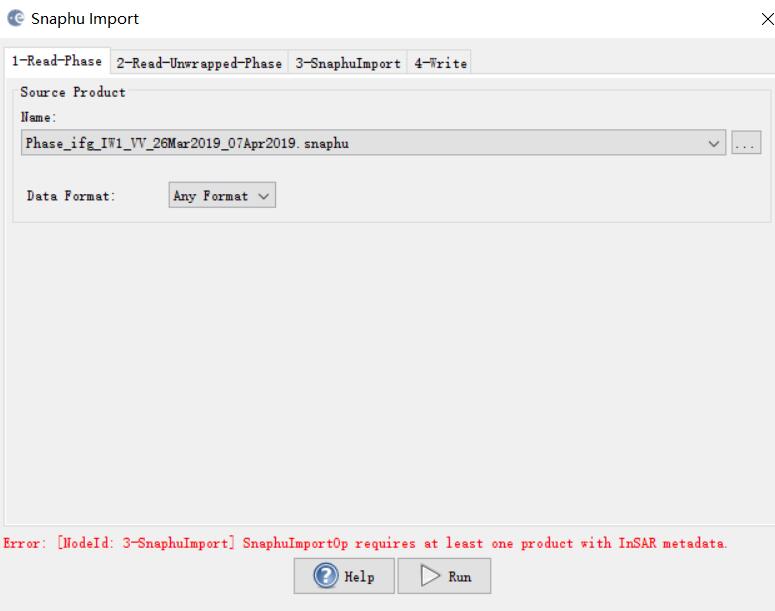
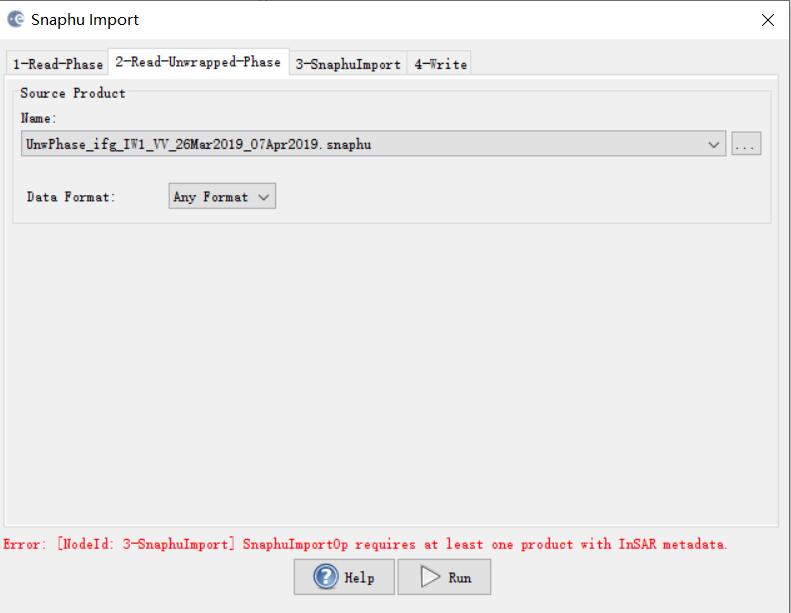
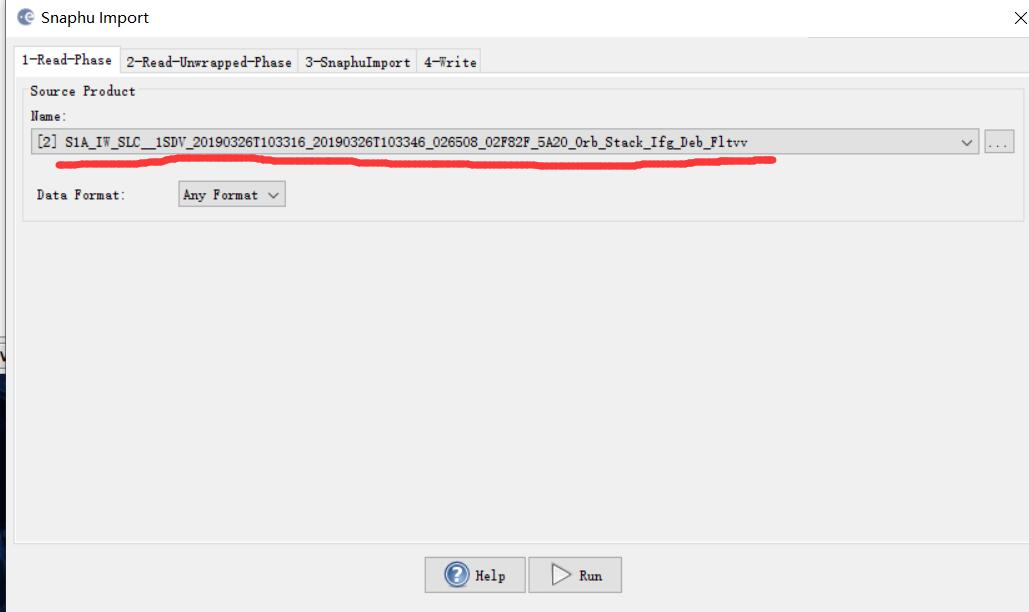
***.dim (1-Read-Phase) is selected, and UnwPhase.snaphu.hdr (2-Read-Unwrapped-Phase) created by snaphu is selected. The problem (snaphuImport requires at least one product with InSAR metadata) is solved. But if Phase_ifg.snaphu.hdr (1-Read-Phase) is selected instead of ***.dim, there will be the error.
In the 1-ReadPhase tab, you should chose the inteferogram before unwrapping which is opened in SNAP by selecting it from the dropdown menu (not over the browse button).
Hello,
I do have the same problem about the height error. Checking the metadata, I got the following values:
- unwrappedPhase.hdr:
– lines: 9172
– samples: 5000 - interferogram, band_IW1_HH (which was used for snaphu):
– num_output_lines: 15060
– num_samples_per_line: 22122
Do I somehow need to downsample the interferogram? How would I do that?
And why would the two images be so different in size in the first place?
(tried to import the files both in both orders, with and without opening them in SNAP before, etc.)
the product you select in the snaphu export determines the extent and dimensions of the unwrapped phase.
Please compare if the hdr of the exported files (phase and unw_phase) show the sames samples and lines.
If so, you probably selected the wrong input during the export (maybe one was multi-looked and the other wasn’t). Remove all products from SNAP and only load the interferogram you want to unwrap and select it in the export.
yes I do that
.
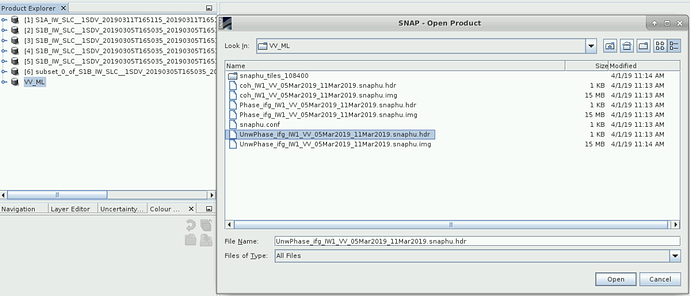
Hello sir, I’m also experiencing the same problem. I followed your tutorial and I also watched a couple of videos on YouTube, but I just couldn’t find my way around it. See screenshots
in your first screenshot, there is no img of the unwrapped interferogram.
Yes. but i have unwrapped the interferogram, I don’t know why it is not coming up. It is surprising.
SNAP needs it i exaclty this folder and with the same file name as the hdr file. Otherwise the import will fail.
Yes. what do you think I am probably doing wrong. I have been on it since yesterday.
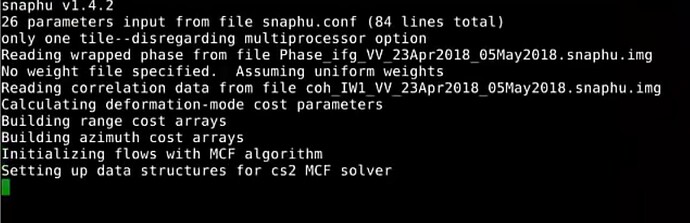
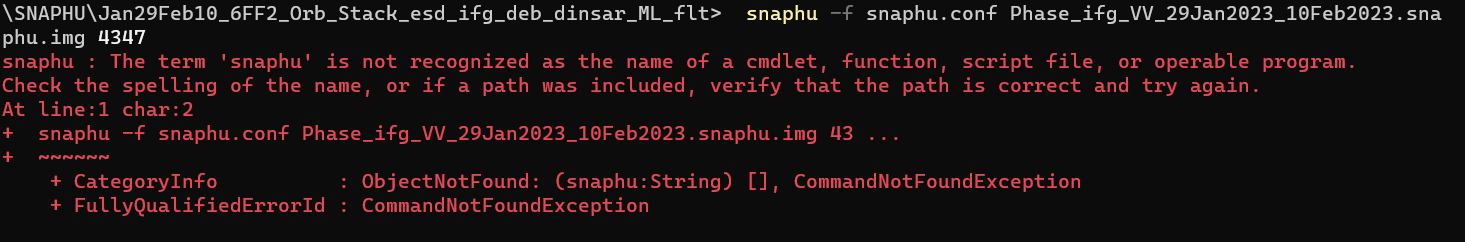
probably snaphu did not finish the unwrapping correctly. Did you get any message of completion? Did you run it inside SNAP or in the command line?