hello guys i am new to this data any of know about deriving chlorophyll, turbidity from sentinel 2data with 10 m resolution .
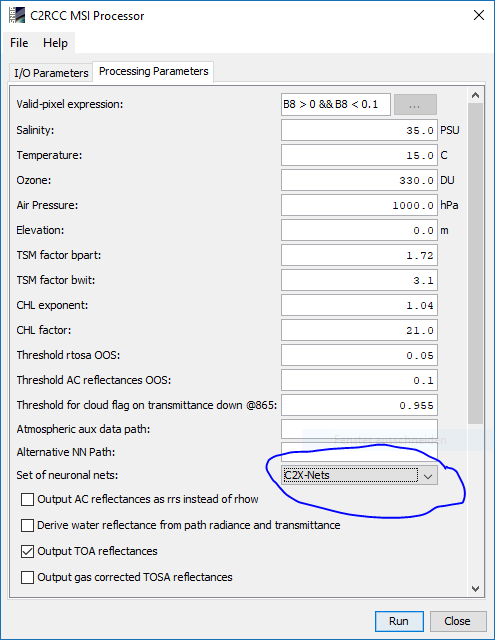
You can do it with the C2RCC processor. You fin it in the menu at Optical \ Thematic Water Processing \ C2RCC Processor\ S2-MSI. You need to resample to 10m before hand.
Thank you for ur kind reply…and from reg the sen2 data in my region L1 c data only available …if u don’t mind if u have any procedure for retrieve chlorophyll please send me sir…
I don’t know if I understand your question.
You need to get the L1C data and then process them with C2RCC. In the output you will have Chlorophyll and TSM. You can get the L1C data from the sciHub (https://scihub.copernicus.eu/)
yes thank you…I did C2RCC for my area then i saved my product .In my product which contain CHL and TSM concentration its shows value but only thing is i found much value around CHL 149 mg/m3 and TSM 56 which concentration are not possible in my study area . may i know solution for getting better estimate
You can try the other neural net. It is selectable in the UI.
It is made for region with high TSM
You can also try to adjust salinity and Temperatur to your area. But in general this has only a minor influence on the results.
Hello,
Is it necessary to do “sen2cor” before resampling the data?
Which is the correct procedure:
- Sen2cor - S2 resampling processor - C2RCC processor
- S2 resampling processor - C2RCC processor
- the other?
You should not do sen2cor. C2RCC needs the L1C data. It does its own atmospheric correction.
sir you mentioned above Neuronal net …may i know which methods is best for estuary region
Thank you very much, Sir.
As estuaries tend to have high TSM I would suggest to use the C2X neural net.
for chlorophyll any neural is there ? for both CHL and TSM after C2RCC process is further band math is require?
The neural net is not only for chl, but for everything the processor generates.
The processor directly generates the chl and tsm concentrations. The bands are named conc_chl and conc_tsm.
If you want to learn more about the processor you can have a look at this document. It is also linked from the user manual in SNAP Desktop.
I tried . for me i adjust temperature and salinity (26 34) in C2RRC and i selected C2X neural . but it processing takes too long around 15hrs…and above mentioned temp , salinity is average of study area shall i enter that value … may i know usually this process how much time will take ?
Have you used the S2 resampling? Try the generic one (Raster / Geometric Operations / Resampling). I guess it will be faster.
To which resolution have your resampled? You can try a coarser resolution.
If you don’t need the uncertainties in the output, disable their computation. This takes also a fair amount of time.
I just did a computation with the generic resampling (to 10 meter) and without uncertainties. The processing finished in less than 70 minutes. The scene was roughly 50% covered by land. My computer is a Intel i7 5th generation 16 GB RAM. Nothing fancy.
The average temperature and salinity value for the area is okay. Maybe adapt them to the season. But as I said earlier, the effect is of minor order.
okay sir i will do as per ur instructions thank you for your kind reply
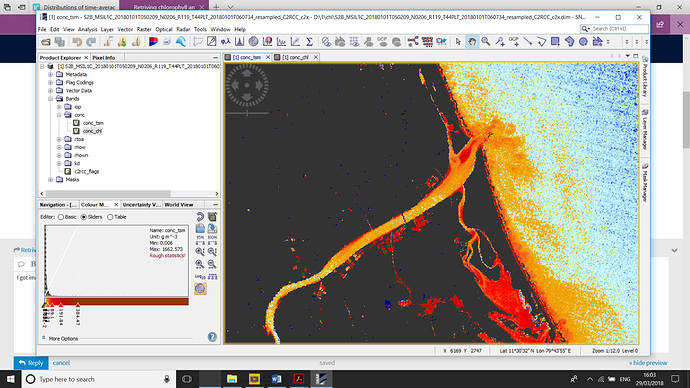
I got image like this . TSM
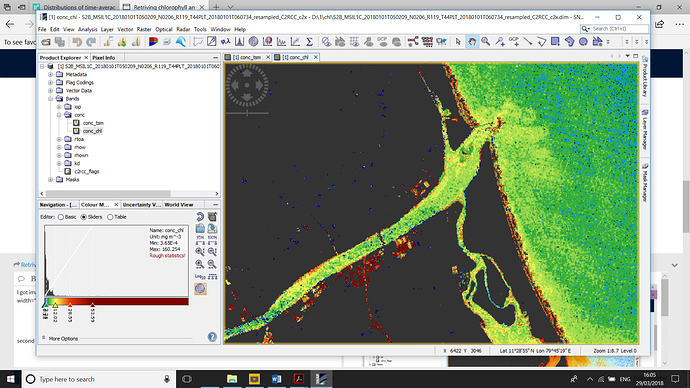
second image chl
were the both image tsm and chl concentration are too high i did C2X neural …is the image is correct or not ? and i have some doubt in both image
is any of these parameter (CHL,TSM) is possible to over estimate ?
why i am asking this in that image coast line are having high values
After processing of C2X is possible to convert data format like (geotiff or any format ) I tried covert format to geotiff but it says about limit file size
I saw some algorithms for retriving chlorophyll but they did for S3 OLCI band and that paper have some tsm algorithm R665/R560
for CDOM R665/R490 , R665/R560
for above band are i could visible in C2X final data is possible to get CHL,TSM value by apply band math ?
For sentinel 2 L1C product which resample process is suitable for retriving Chlorophyll and TSM In SNAP i could find two option
1-Raster-Geometric operation-Resampling
2-Optical-Geometric-S2 Resample processe
what’s diference between above two tools…
Thank You
There is a significant difference in the resampling of angles layers. When using the C2RCC algorithm I would suggest to use the S2 Resampling Processor.
A quick look of the differences is reported here:
yes i could see the differences … once again Thank You …