Another request/idea for the SEN2COR python script: Currently, the original *.jp2 tiles are georeferenced, the corrected tiles aren’t. It is easy to add the georeferencing manually, but in my opinion, it makes sense if this would be incorporated in SEN2COR. Currently I’m using gdalcopyproj.py which actually works great.
Hello MWess.
The issue you were facing manifests only if using GTK+ look and feel.
I have tracked down this issue to be caused by an improper window hierarchy and I have corrected it.
But until an update of the module is published, you can switch to another look and feel (from Tools > Options menu) - “metal” for example.
Hi @ all,
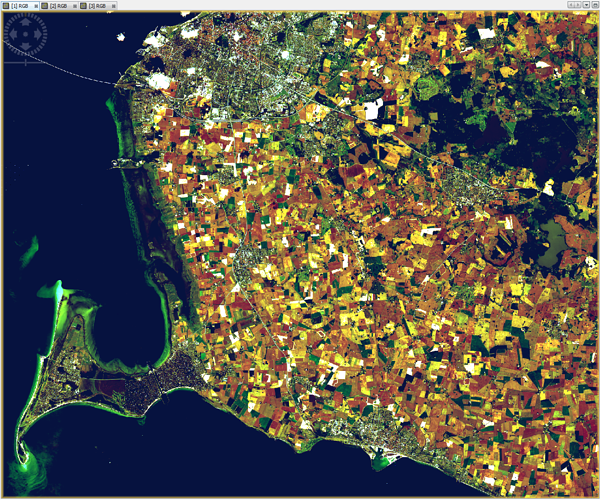
i have installed sen2cor 2.0.4 and processed L2A images in 60m, 20m and 10m resolution. The 60m and 20m L2A images are looking good, but there is something wrong with the 10m resolution, see RGB images attached. The DNs of 20m and 60m are very similar, but different in 10m. For example, on grassland I have in channels b02, b03, b04, b08 DNs of 20, 60, 10, 100 in 20m and 60m resolution, but in 10m resolution on the same grassland there are DNs 8, 26, 8, 525.
Any idea?
Regards,
Ralf
60m
20m
10m
Dear all,
I have successfully processed an L1C dataset using the sen2cor cmd line. The L2A_Process was run with the --sc_only parameter since I am mostly interested in scene classification. I opened the resulting dataset in SNAP and was wondering how to visualize the scene classification? i.e. how do you arrive at the visualization shown in the attached?
With Thanks!
Use the Long Path Tool. It is really nice tool.
Use the Long Path Tool. It is really nice one.
The scene classification is not supported by the actual version of the Sentinel-2 Toolbox. We’ll work on it to propose it as soon as possible.
Hi Ralf,
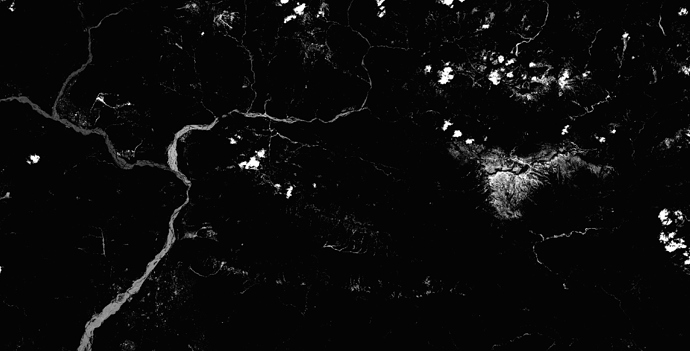
I have the same problem:
B02 (Blue) after SEN2COR in 10m:
B02 (Blue) after SEN2COR in 20m:
Is this a mistake in the SEN2COR script or is there again some scaling problem in the data?
EDIT1:
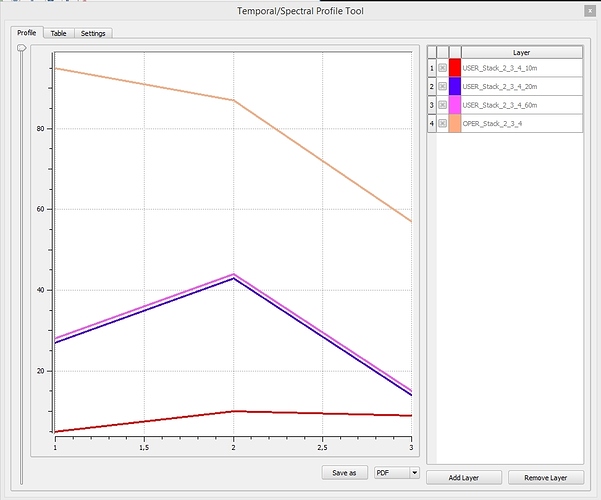
Here a comparison of the spectral profiles of the bands 2, 3 and 4 in 10m-resolution before the SEN2COR-Correction, and then in 10, 20 and 60m resolution AFTER the atmospheric correction. Shouldn’t the 10, 20 and 60m-images be similar? I picked a forest Pixel in a homogenous region. The 10m result seems to be really off!
BR
Michael
EDIT2: Another Question: Is there a reason why for the corrected dataset, Band 08 is only available in 10 m, where all other 10m bands are also written in 20 and 60m?
Hi,
Can you already tell when the issues with the sen2cor toolbox will be resolved?
Currently, the most “interesting” resolution (10m) is not usable after applying the atmospheric correction.
BR
Michael
dear Michael and Ralf, The 10m problem you observed is analyzed, please read my statement just published on the Sen2Cor status.
dear Jochem, the 10m problem you observed is analyzed, please read my statement just published on the Sen2Cor status.
Hi,
I am getting only the post template files in my working directory. I am still unable to get L2A data from sen2corr each time I run it I get this snap error message. Any idea please. Thanks
Hello,
i am trying to run Sen2cor (2.0.5) in SNAP (2.0-beta-08) as an external Tool.
I installed sen2cor sucessfully and the L2A_Processor is working on command line level.
If you need, i can provide my command line output from L2A_Processor.
I am working on a Windows 7 64bit machine.
I followed the discussion in the forum, but i am still having problems to create a L2A through the integrated sen2cor Tool in SNAP. Heres what i have done so far:
- I renamed the root folder to:
S2A_OPER_PRD_MSI_L1C_to get rid of path length issues - Local Input path of the XML:
D:\beck_ct\data\s2a\L1C\S2A_OPER_PRD_MSI_L1C_
I keep getting following error:
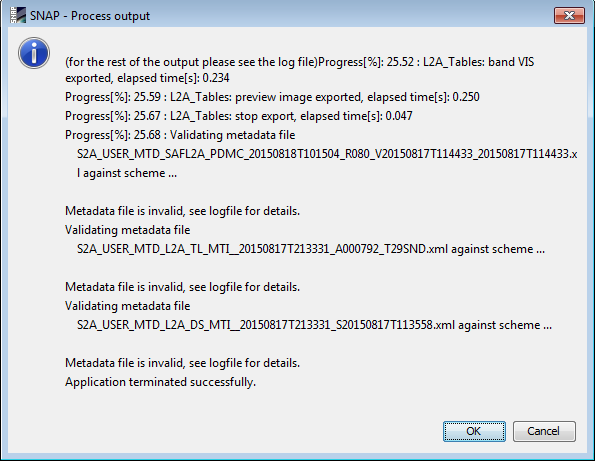
Is it possible that those errors are produced through the renaming oft the root folder?
Many Thanks in Advance,
Christian
Dear Christian,
You get this message because you chose to see the processor output. There is a message saying Metadata file is invalid because of compliance to PSD (the product spec) but ‘Application terminated successfully’ so apparently everything went fine. What you see is a warning message that is not related to the folder.
Dear Nicolas,
thanks for your quick reply.
So for now we dont have worry too much about the metadata Validation errors?
Hello @chris,
For the moment, no. @umwilm will tell you more about it.
Please notice that a new release of sen2cor (2.0.6) is now available, with a few important fixes. Be sure to update !
Hi @sgri0009 !
Hope you noticed that now with the latest SNAP 2.0, the scene classification data is read automatically.
It appears in the “Index Bands” category, and tuning the color manipulation can quickly get you to the visualization you want.
Also, for each class, there is a dedicated Mask that can be used in SNAP, so you can visualize each class independently.
Hi,
The validation warning (error) currently appears due to a “mixture” of V12 and V13 in the L1C input data. This will be cleaned up in the coming version, as we currently received a new V13 Metadata scheme which must be integrated.
it does not have impacts on the generated output product.
Hi,
thanks for clarification @umwilm!
@jmalik: I recognized the new version of sen2cor you provided. Thanks for the advice!
General procedure for updating to a new version? I propose run install.py?
Thank you,
Christian
Great!
downloaded SNAP 2.0 and will give it a try.
Thanks