Hi @fschlenz
It is a bit hard to explain, but as I understand it the Python version is a complete recoding of the Fmask paper and therefore does not use any of the Fmask author’s original Matlab code, the Matlab version for Sentinel is simply the Landsat version made compatible with Sentinel. So the Matlab version is closer to the original.
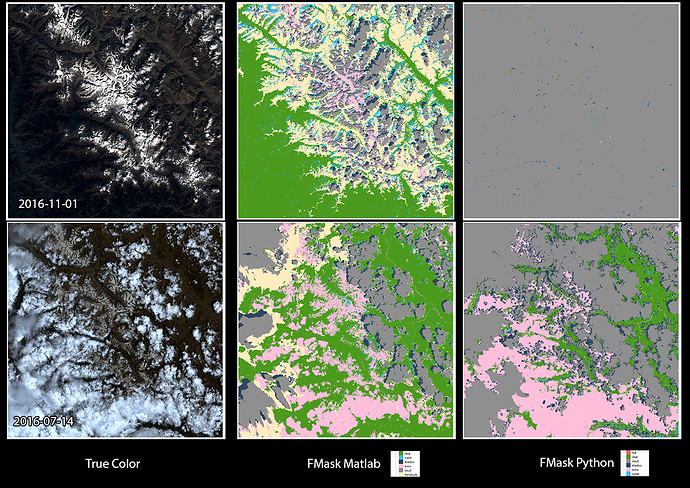
But a couple things I notice, the Python version takes significantly longer 1-2 hours (at 10m resolution) vs 2-3 minutes of Matlab (note: the Matlab version is always 20m resolution). I haven’t tested Python at lower resolutions. In some extreme cases the Python version for my data marks the image as 100% cloud cover, when the Matlab version is around 30%… So that is a clear error.
The data I am using to test this data is not ideal, but that was my intention. (North India)
Here is a image for comparison… you can see that neither are perfect, but Fmask matlab seems more consistent.
Hope that helps.
As for the Matlab code change, in reply to @unnic
In the file Build_SunViewGeometry_MSI.m around lines 155 to 200 or so, you need to alternate between the following code depending on if it is before or after August 2016, but this seems to work for most of my data.
Pre-August:
%%%%%% PRE-AUGUST
if length(S)==1
DetectFootPrint.Ndect(1) = str2num(S(1).eop_colon_MaskFeature.Attributes.gml_colon_id(24:25));
disp('attention')
DetectFootPrint.Nband(1) = find(strcmp(bandIdList, S(1).eop_colon_MaskFeature.Attributes.gml_colon_id(21:22)));
Vect = str2num(S(1).eop_colon_MaskFeature.eop_colon_extentOf.gml_colon_Polygon.gml_colon_exterior.gml_colon_LinearRing.gml_colon_posList.Text);
Vect = reshape(Vect' , [3 length(Vect)/3 ])';
DetectFootPrint.Data{1,1} = Vect ;
% Z = MSIinfo.Angles.VZA(:,:,MSIinfo.Angles.bandIdList==DetectFootPrint.Nband(1),MSIinfo.Angles.DeteIdList==DetectFootPrint.Ndect(1)) ;
else
if Pppplot == 1
figure
hold on
sym = 'oxoxoxoxox';
end
for i = 1:length(S)
DetectFootPrint.Ndect(i,1) = str2num(S{i}.eop_colon_MaskFeature.Attributes.gml_colon_id(24:25));
% DetectFootPrint.Nband(i,1) = strcmp(MSIinfo.bandIdList , S{i}.eop_colon_MaskFeature.Attributes.gml_colon_id(21:22));
DetectFootPrint.Nband(i,1) = strcmp(bandIdList , S{i}.eop_colon_MaskFeature.Attributes.gml_colon_id(21:22));
Vect = str2num(S{i}.eop_colon_MaskFeature.eop_colon_extentOf.gml_colon_Polygon.gml_colon_exterior.gml_colon_LinearRing.gml_colon_posList.Text);
Vect = reshape(Vect' , [3 length(Vect)/3 ])';
DetectFootPrint.Data{i,1} = Vect ;
if Pppplot == 1
IDX = knnsearch(X10m,DetectFootPrint.Data{i,1}(:,1));
Ximg = X10m(IDX);
IDX = knnsearch(Y10m,DetectFootPrint.Data{i,1}(:,2));
Yimg = Y10m(IDX);
plot(Ximg,Yimg,'k')
Z = MSIinfo.Angles.VAA(:,:,MSIinfo.Angles.bandIdList==DetectFootPrint.Nband(i),MSIinfo.Angles.DeteIdList==DetectFootPrint.Ndect(i)) ;
scatter(X5kmat(:),Y5kmat(:),30,Myreshape(Z),sym(i))
end
end
end
% axis image
%%%%%% END PRE-AUGUST
Post-August:
%%%%%% POST-AUGUST
if length(S.eop_colon_MaskFeature)==1
DetectFootPrint.Ndect(1) = str2num(S.eop_colon_MaskFeature.Attributes.gml_colon_id(24:25));
disp('attention')
DetectFootPrint.Nband(1) = find(strcmp(bandIdList, S.eop_colon_MaskFeature.Attributes.gml_colon_id(21:22)));
Vect = str2num(S(1).eop_colon_MaskFeature.eop_colon_extentOf.gml_colon_Polygon.gml_colon_exterior.gml_colon_LinearRing.gml_colon_posList.Text);
Vect = reshape(Vect' , [3 length(Vect)/3 ])';
DetectFootPrint.Data{1,1} = Vect ;
% Z = MSIinfo.Angles.VZA(:,:,MSIinfo.Angles.bandIdList==DetectFootPrint.Nband(1),MSIinfo.Angles.DeteIdList==DetectFootPrint.Ndect(1)) ;
else
if Pppplot == 1
figure
hold on
sym = 'oxoxoxoxox';
end
for i = 1:length(S.eop_colon_MaskFeature)
DetectFootPrint.Ndect(i,1) = str2num(S.eop_colon_MaskFeature{i}.Attributes.gml_colon_id(24:25));
% DetectFootPrint.Nband(i,1) = strcmp(MSIinfo.bandIdList , S{i}.eop_colon_MaskFeature.Attributes.gml_colon_id(21:22));
DetectFootPrint.Nband(i,1) = strcmp(bandIdList , S.eop_colon_MaskFeature{i}.Attributes.gml_colon_id(21:22));
Vect = str2num(S.eop_colon_MaskFeature{i}.eop_colon_extentOf.gml_colon_Polygon.gml_colon_exterior.gml_colon_LinearRing.gml_colon_posList.Text);
Vect = reshape(Vect' , [3 length(Vect)/3 ])';
DetectFootPrint.Data{i,1} = Vect ;
if Pppplot == 1
IDX = knnsearch(X10m,DetectFootPrint.Data{i,1}(:,1));
Ximg = X10m(IDX);
IDX = knnsearch(Y10m,DetectFootPrint.Data{i,1}(:,2));
Yimg = Y10m(IDX);
plot(Ximg,Yimg,'k')
Z = MSIinfo.Angles.VAA(:,:,MSIinfo.Angles.bandIdList==DetectFootPrint.Nband(i),MSIinfo.Angles.DeteIdList==DetectFootPrint.Ndect(i)) ;
scatter(X5kmat(:),Y5kmat(:),30,Myreshape(Z),sym(i))
end
end
end
%%%%%%%% END POST-AUGUST
The main cause of the error was the way the directory/struct in Matlab was read, for example, in pre-August it might have been 1 folder with 12 files, post-August it turned into 12 folders with 1 file each… or something like that.
Cheers

 )
)