Hi Everyone,
I am processing S1 SLC images in python, unfortunately I have to use snappy to calibrate the images in Python before I can use them. I run my script to process multiple SLC images
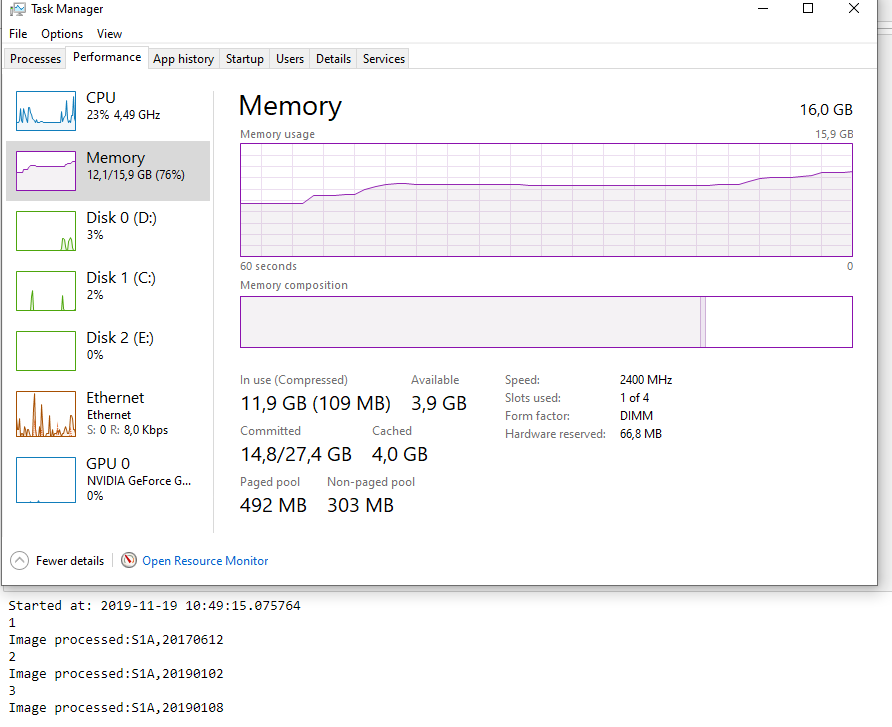
I am not exactly sure how it works, but it seems to me there is some sort of issue with the allocation and clearing of memory in snappy.When I look at the memory usage in windows, I notice that with every image processed, the Standby (cached) memory increases. This happens specifically in the part of the script where I am using the snappy module to Calibrate the SLC images.
This memory is not cleared or released, even though the Python script started processing the next SLC image. I tried several tricks in Python (garbage collecting, multiprocessing, etc…) but it wont seem to clear this cached memory.
Could anyone familiar with snappy help me with this issue… So far it has been driving me nuts…
This is the part of the code responsible for reading and calibrating the SLC images:
print('Reading...') time_1 = datetime.datetime.utcnow() # ~~~~~input data~~~~~ tl_x = 0 width = meta['samplesPerBurst']
tl_y = 0 height = meta['linesPerBurst'] * len(burst['azimuthTime'])
# ~~~~~Snappy Calibration~~~~~
GPF.getDefaultInstance().getOperatorSpiRegistry().loadOperatorSpis() satdata = ProductIO.readProduct(loc)
# Calibration parameters = HashMap() parameters.put('outputSigmaBand', True) parameters.put('selectedPolarisations', 'VV') parameters.put('outputImageInComplex', True) product = GPF.createProduct("Calibration", parameters, satdata)
print(datetime.datetime.utcnow()) # read bands bi_VV = product.getBand('i_' + swath + '_VV') bq_VV = product.getBand('q_' + swath + '_VV')
# create numpy variables i_VV = np.zeros((height, width), dtype=np.float32) q_VV = np.zeros((height, width), dtype=np.float32)
i_array = bi_VV.readPixels(tl_x, tl_y, width, height, i_VV) q_array = bq_VV.readPixels(tl_x, tl_y, width, height, q_VV)
slc = i_array + 1j * q_array
# delete product from memory satdata.dispose() product.dispose()
import jpy System = jpy.get_type('java.lang.System') System.gc()