could you please also show us the ProductSet Reader Tab?
looks the same as in my case.
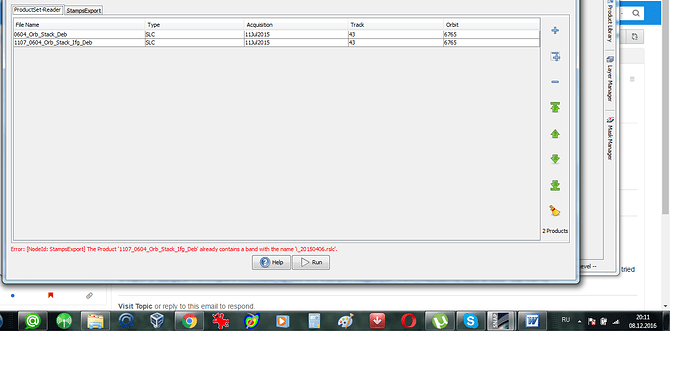
Can you please also show a screenshot of all the bands in the two products?
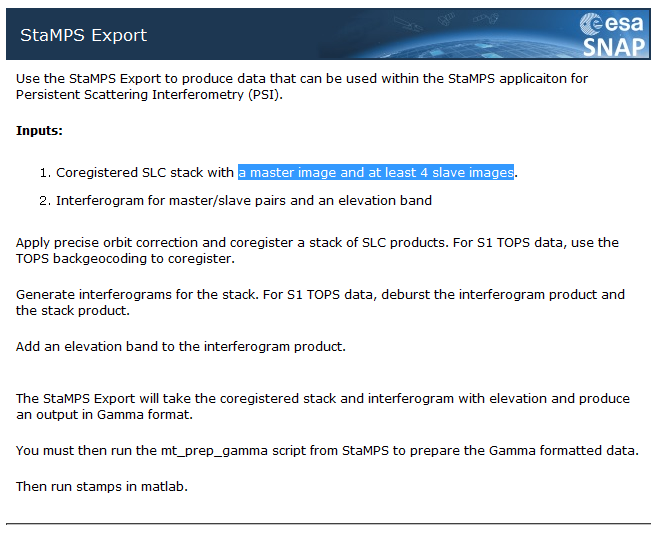
according to the Help menu your stack must have at least 5 SLCs (1 master and 4 slave bands)
The rest looks like my data.
Dear ABraun,
Thank you for your time!
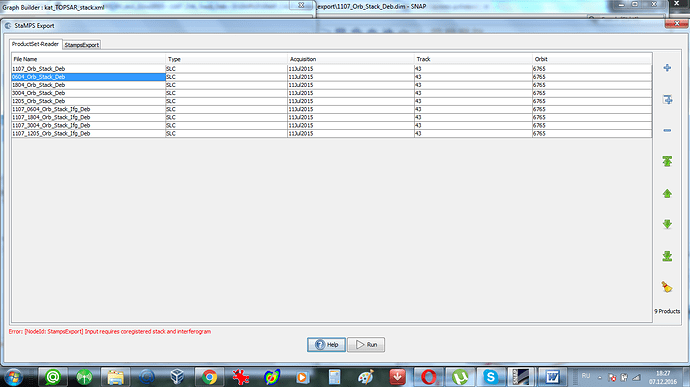
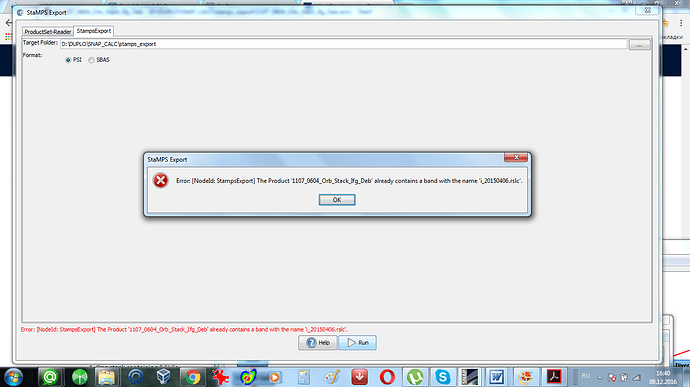
Yes,4 ifgs and 5 SLCs - it is the requirement of Stamps. This is necessary for mt_prep_gamma. And at first I tried 5 SLCs with corresponding 4 ifgs.But in this case “Stamps export” from SNAP did not work at all. I had the error which I put in my earlier post (I attached it once again). Luis Iveci wrote:
“I could update the stamps export to allow multiple individual interferograms. This would allow you to choose slave-slave pairs”.
I suspected that probably at present pairs (1 ifg and 1resampled image) can be exported to Stamps only individualy… But it is only my presumption…
If somebody from the community have already used the SNAP “export to Stamps” with S-1 images successfully , please, share your experience!
Katherine
Dear Katherine,
At first, you should co-register your images using back-geocoding functionality, that supports Sentinel stack co-registration now. (Radar -> Coregistration -> S1 TOPS Coregistration -> S-1 Back Geocoding)
Secondly, SNAP didn’t allow me to export with “topo-phase removal” step done. So I didn’t do it by now.
Mine processing chain looks like this:
Apply Orbit File to each image -> S1 tops split -> back geocoding of stack -> interferogram formation -> Add elevation band to last product in the chain
Talking about deburst and subset operations I am not fully sure yet. It seems that you can deburst, subset co-registered stack and interferogram stack individually or deburst, subset at first and then generate interferograms out of subset.
Don’t exprort too small area. It should be definately larger than 2km x 2km. Otherwise StaMPS will find zero permanent scatterers.
During step nr. 6 you may need to decrease values of parameters “unwrap_grid_size” or “unwrap_gold_n_win”. The less image you exort, the more parameters will need to be decreased.
I still have some problems incide StaMPS though. The image is stretched, points are geocoded near zero co-ordinates and deformation values are unnaturelly huge.
Jaroslav
Dear Jaroslav,
Thank you very very much for sharing your experience!It will save me a lot of time. I will try to follow your advice and inform you about the results. I will be happy to share my experience if I succeed.
Thank you again!
Katherine
Dear Jaroslav,
Sorry to disturb you but one more question. How did you choose master in the new back-geocoding functionality, that supports Sentinel stack co-registration now. (Radar -> Coregistration -> S1 TOPS Coregistration -> S-1 Back Geocoding)?
Thank you for your time.
Katherine
[quote=“katherine, post:30, topic:3211”]
uestion. How did
[/quote] Sorry, master image should be just the first in the ProductSet-Reader of S-1Back-Geocoding
Dear Luis and all the colleagues,
At first I I wish to thank all that have labored to bring this into being! I have exported S-1 data from SNAP to Stamps and Stamps worked from the beginning to the end! Having the possibility to work with PSI makes SNAP much more powerful tool! It is really great! But there are few things which prevent using this wonderful tool at present. I do not know how to fix them without your assistance.
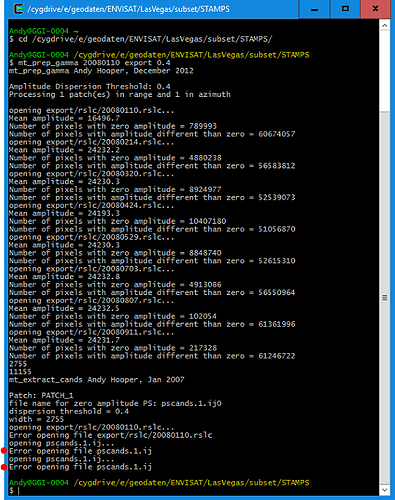
1/ Stamps needs ifgs with subtracted reference dem (srd). “Stamps export” from Snap refuses to export ifgs after TopoPhaseRemoval. Only ifgs after functionality “Interferogram formation” (only flat Earth subtracted) can be exported. Below is the error message which I get in the case of exporting results of TopoPhaseRemoval.
I tried just to copy bands from the Product after TopoPhaseRemoval to the Product after Interferogram formation. But in this case only /geo and /rslc directories were created (no /diff).
2/ In the /geo directory there should be files YYYYMMDD.lon, containing one float32 value per pixel in master crop, and same for lat. The mt_prep_gamma script will extract lon/lat from binary raster files. Without this StaMPS can not do PS geocoding and plot any results.
3/ I got some strange values after “Stamps export”. In the .par files for ascending orbit I have Heading = 105.94474020664796 degrees (instead of -13.84 according to “annotation”) lambda=55465.8 (what are the units?) In Stamps lambda is used to be in m, so for S-1 probably it should be 0.0554658?
Thank you again for all your hard work and looking forward to have solutions to overcome these problems.
All the best,
Katherine
Dear Katherine,
great to hear that you are making progress. I’d also like to use StaMPS and have a few questions:
- Are you working on a Linux system?
- Which was the first step (in the whole StaMP
PS workflow) you performed after the SNAP export? - Which version of StaMPS are you using?
Maybe then I can help you testing the SNAP export and we can compare outcomes regarding possible error sources.
Thank you also for your valuable comments which might help to improve the software.
Dear Jaroslav,
Thank you again for all your advice. I launched Stamps succesfully! I think that the problems incide StaMPS are due to the absence of lon lat files in /geo girectory. See my post below.
All the best,
Katherine
Dear Andreas,
I will be very glad if we manage to overcome problems of SNAP to StaMPS export together. It is really very usefull SNAP’s funcionality. I will be very glad to share my experince in StaMPS. Answering your questions.
1/ Yes, I work on Linux 14.04. Stamps works only with Linux. But it is also possible to do all SNAP work on Windows machine (including export to Stamps) and then just copy the whole directory with exported files to a Linux machine and continue there.
2/ The first script to launch is mt_prep_gamma (bin directory in Stamps).
From your working directory (the directory where you have /rsls, /geo, /diff directories fron SNAP):
$mt_prep_gamma /full path to your working directory/ 0.4
(0.4 - amplitude dispersion threshold for initial choosing of PS). This is in case your area is quite small and you plan to have one patch. My area is about 7х7 km and one patch appeared to be OK. This script creates PATCH1 directory in your working directory . Then in the working directory you launch matlab and in matlab run stamps. For example like this
stamps (1,6).
3/ I use the latest version of StaMPS_v3.3b1.
If you have any other questions about StaMPS, please, ask.
Good luck
Katherine
thanks for your reply.
Some of the StaMPS scripts also worked for me on windows, I was able to compile it using cygwin. However, non of the mt_prep scripts works on both Linux and windows. There’s something with the coding of the data because parantheses seem to cause errors. I will have a closer look at it.
Dear Andreaus,
Below is the answer of Andy Hooper (the author of StaMPS) on the MAINSAR forum about using Stamps under cygwin. Probably this will save your time.
Re: [MAINSAR] Re: Problems of StaMPS
Перевести сообщение на русский
Hi Annie,
I have never tried to run StaMPS under cygwin, and I imagine there will be some problems. The easy option would be to add linux to your windows machine (dual-boot).
Andy
All the best
Katherine
thanks for clarification.
I get the same error on Linux and on Windows. So I can exclude the operating system as an error source, at least for this case.
Could you please clerify what error do you have and at what step?
Katherine
nevermind, found the mistake.
I’ll try the further steps and will tell you as soon as possible.