Dear soukainamez,
I don’t know what is your matlabs problem, but my experience is that It is important to use at least 7 interferograms to get a selection of points that will not have any problems with matlab (in particular my problems beginning during step 3).
Good luck.
thank you annamaria for your reply
Hello everyone,
Did someone have some experience about SBAS processing (using STaMPS) of interferograms (from SNAP).
Thanks in advance for any advice.
Hi kleok,
Unfortunately, at present SNAP does not support export to StaMPS for StaMPS-SBAS processing. Hopefully this will be done in future releases.
Katherine
Yes that is the plan.
Hi @juanesburgo
Did you get rid of your problem with PS on water, i have the same problem her :
please if there is a solution let me know.
Dear All,
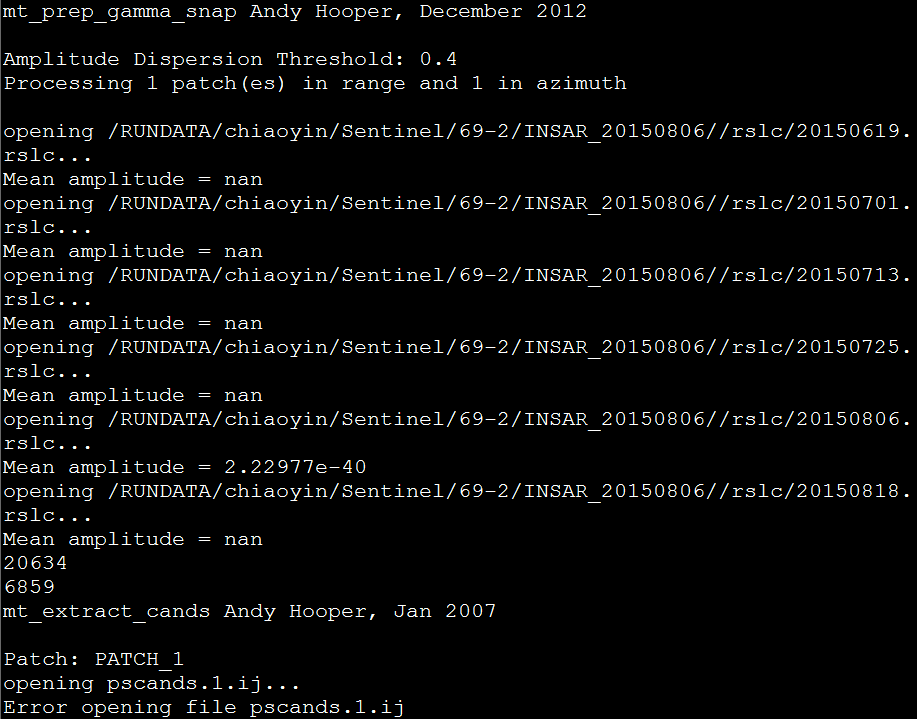
After running make install from src directory, I got this message, is that mean my installation is correct ??
After that when I am running mt_prep_gamma_snap then I am getting this message,
Although, I have some output in the working directory,
CAN ANYONE PLEASE TELL ME IF I AM IN THE RIGHT WAY OR WHAT IS GOING WRONG:::
Thank you in advance…
@katherine @ABraun @lveci @Abdel @annamaria @falahfakhri @FeiLiu @Sharon @Adrian
Bayzidul
Have you found any other alternative way for SBAS processing? Thanks
Hi everyone. First of all, many thanks to you for your kindly help for all questions.
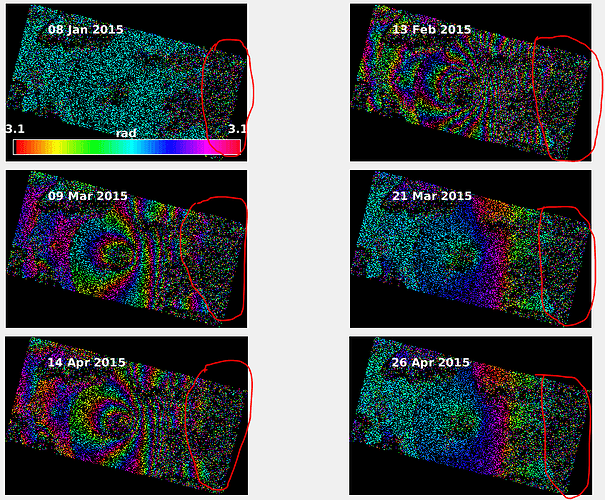
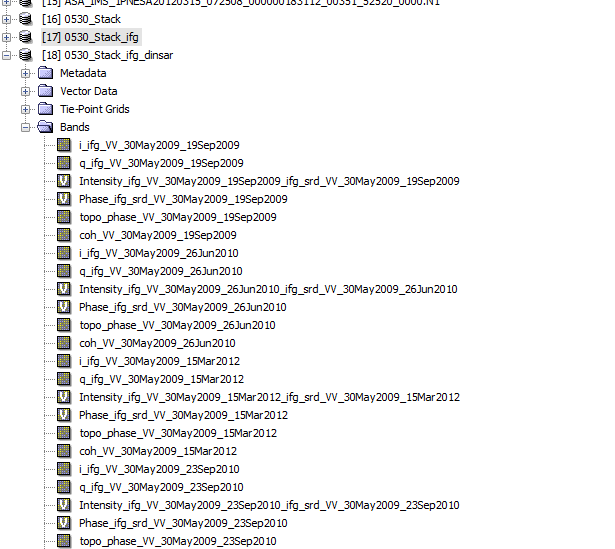
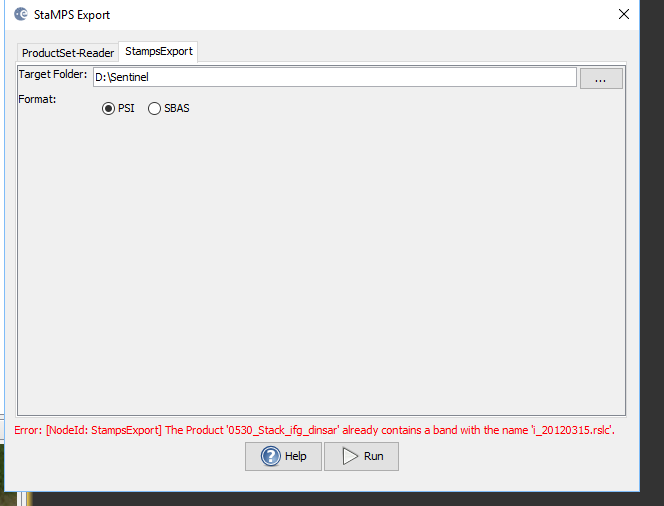
My problem is about StaMPS export. I am using 15 Envisat IMS_1P SLC data. One of them is Master and the others are Slave. First, I coregistered these data succesfully, then I applied Interferomgram Formation and after Topographic Phase Remove. I followed instructions which are I picked up from this forum. But I got an error, could you give me an advice to fix it? Thanks.
I personally would try to remove all name parts starting with _2009_ifg_srd_, then save the product an run again.
I fixed the error by deleting the same name products. I have two 2012_03_15 image with different path. Then I deleted one of them, PSI export processing started. Thanks.
I am using GMTSAR for interferometric processing and SBAS processing. Also, i am trying to import the generated unwrapped interferograms from GMTSAR into GIAnT software package in order to compare the different SBAS results.
Hi @bayzidul I finally achieved to export StaMPS .dem. rslc. .geo and .diff folders and I installed StaMPS same with you and got same messages. But I do not know how should I do first? I have to do all chapters in manual or could you give me an advice? For example how can you got PATCH.X files. I guess I need to get PATCH.X files then I will run MATLAB stamps.m ? Is it true? Thank you.
Hi @Fikretjfm
Honestly, I have tried all possible ways to do it but my problem remains same for both platform CYGWIN and LINUX (see my last message with screenshot). If you could run the first script (mt_perp_gamma_snap or mt_prep_gamma_snap(changed) by @FeiLiu ) then you should have PATCH file and other files. Probably @AranLarra @falahfakhri @FeiLiu @annamaria @katherine or someone else could provide us a screenshot of the terminal with code how they are running the first script and also a screenshot of the folder (containing all the file) after running. Then we have to run the second script in Matlab.
If you read previous sms (or the word documnet by @falahfakhri ) then you will understand all situations and you could see that TopoPhaseRemoval has problem with Stack processing. Therefore, I am now doing everything manually as mentioned @AranLarra and only then I can let you know my status by the weekend.
Cheers
Bayzidul
@bayzidul ps_load_initial_gamma_snap.m and mt_prep_gamma_snap must be in StaMPS installation directory not in your working directory.
In particular:
mt_prep_gamma_snap must be in /bin,
ps_load_initial_gamma_snap.m must be in /matlab.
Dear Annamaria,
I added your last notes to the document I have created, so would you please to participate in this until to reach the complete method and steps, then we could add something concerning the installation of StaMPS under cygwin or linux.
In the same time I’d like to draw the attention of all my colleagues that I’m preparing the steps of SBAS using SNAP, in case someone would like to participate.
With my best wishes,
Falah,
PSI USING StaMPS_V_5.docx (186.5 KB)
I met the same error message in the end of the mt_prep_gamma step:
“Error opening file pscands.1.ij”
I have run mt_prep_gamma inside my “export” folder, but nothing change.
Do you know how to solve this problem?
Thank you very very much!
I don’t have a solution for that, I stopped using Windows with StaMPS because of these errors.
Hi @ABraun
But I used Linux system not Windows for processing. With the version of SNAP is v6.0, and the version of StaMPS is v3.2.1.
And another question, the “mean amplitude = nan” is correct?
Thank you!