Hi,
I am trying to run the Biophysical Processor tool in SNAP 4.0 to calculate fPAR and FCV.
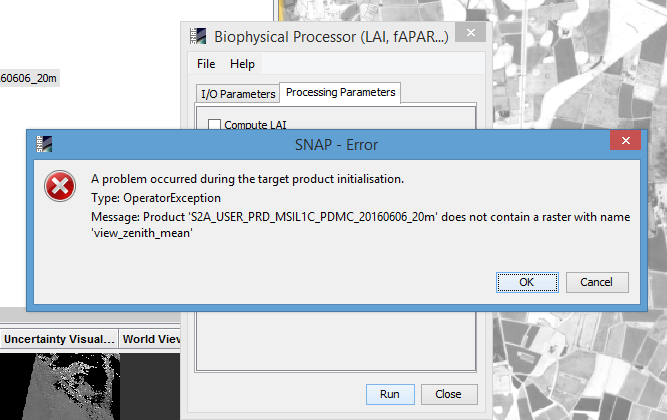
I have first run the sen2cor to obtain BOA L2A product at 20 m and then tried to apply the Biophysical Processor tool to calculate the mentioned products, it gives me the product dose not contain a Raster with view_Zenith_mean ! see image belowplz.
Anyone knows any idea how to solve it ? thanks a lot
Hi @Daniel1
Did you update SNAP recently ? Because this issue is fixed with the latest modules updates.
I have updated SNAP to SNAP 4.0 after removing the old version , but still have this issue!
Hi Julien,
I would like to know if there is any documentation about the Biophysical processor ?
Do you know where i can find this ?
thanks
Hi @insight ,
The ATBD of this processor is available here: http://step.esa.int/docs/extra/ATBD_S2ToolBox_L2B_V1.1.pdf
thank you for your help !
Hi jmalik,
I have the same problem with SNAP 5.0.8.
The product I use is a L2A product from THEIA.
For information, when I check the version of the biophysical, it is written “null”.
Thanks in advance
Hi Nicolas,
all the data about the zenith/azimuth angles for each band are available in the file “SENTINEL2B_20171031-143733-460_L2A_T20PQB_D_V1-4_MTD_ALL”.
The problem is SNAP doesn’t read this file (XML generated by MAJA chain).
Is there a way to provide these data to the BioP processor ?
Thanks
Hi,
please, have a look at
I am working on Biophysical Processor on SNAP 5.0 and 6.0 versions, the tool works fine and it generates temporary output in the window. But, when I try to view it in main window, it does not work. Progress bar keeps on working, but no results and even the temporary outputs are not written to any other format. Is there any solution ?
The biophysical processor can take a long time to compute the output depending on your input (if you have made a subset or not). In SNAP, when you apply a processor, if you do not select the “Save as” option:
generally no computation is made, the computation is only made when the data is required for showing it or writing it to the disk for example. Although you can see the output product in the output explorer, it contains only the information for computing it, and when you try to visualize it, then the computation begins.
Hi Obarrilero,
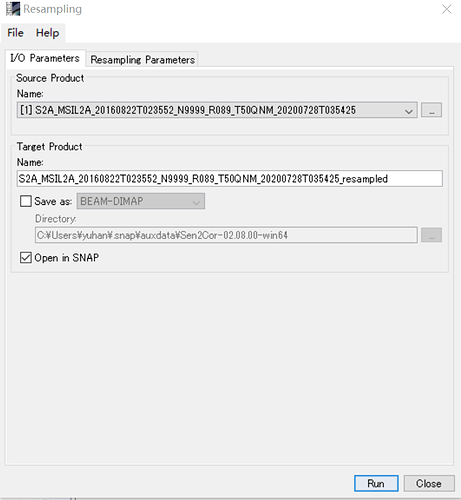
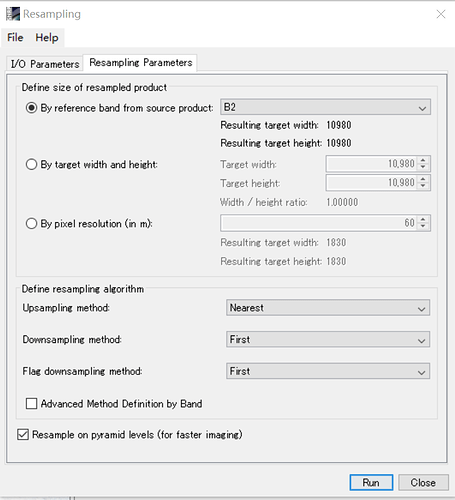
I m working on computing fAPAR from sentinel L2A products in SNAP 7.0. May I ask why we need to resample the bands separately? Can I resample the image into 20m then input it into biophysical processor?
Thanks a lot,
Han
The processer needs the pixel values from different wavelength which are present in different resolutions.
To find the same pixel at the different resolutions the data needs to be resampled.
You have the choice which common resolution you want to resample to. You can choose 20m or 60m or even 10m, or something else.
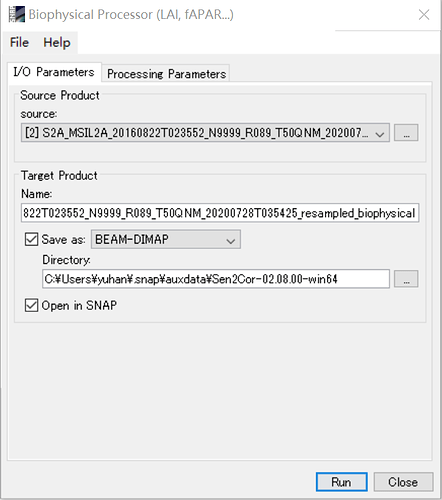
Hi Marpet, Thanks for your reply. I resampled the images to 10 m, but didn’t resample the bands individually. Then I process the images by biophysical processor to compute the LAI and fAPAR as shown in the figures. Does it make sense?
Yes, that’s correct.