I am trying to perform the C2RCC using sentinel 3 that has already been masked and cut using a vector file. I use python and the following code:
import snappy
from snappy import GPF
from snappy import jpy
from snappy import HashMap
from snappy import ProductIO
import os
outFolder=‘F:/hydromerit/Hydromedit/images/sentinel_2/out/20180721_S3_masked_sea.dim’
sal=35.0
temp=15.0
ozo=330.0
pres=1000.0
el=0.0
TSMfakBpart=1.72
TSMfakBwit=3.1
CHLexp=1.04
CHLfak=21.0
thresholdRtosaOOS=0.005
thresholdAcReflecOos=0.1
thresholdCloudTDown865=0.955
outputAsRrs=False
deriveRwFromPathAndTransmittance=False
useEcmwfAuxData=True
outputRtoa=True
outputRtosaGc=False
outputRtosaGcAann=False
outputRpath=False
outputTdown=False
outputTup=False
outputAcReflectance=True
outputRhown=True
outputOos=False
outputKd=True
outputUncertainties=True
OLI=ProductIO.readProduct(‘F:/hydromerit/Hydromedit/images/sentinel_2/out/20180721_S3_masked_sea.dim’)
print ‘C2RCC’
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(‘validPixelExpression’,‘quality_flags.invalid && (!quality_flags.land || quality_flags.fresh_inland_water)’)
parameters.put(‘temperature’,temp)
parameters.put(‘salinity’,sal)
parameters.put(‘ozone’,ozo)
parameters.put(‘press’,pres)
parameters.put(‘TSMfakBpart’,TSMfakBpart)
parameters.put(‘TSMfakBwit’,TSMfakBwit)
parameters.put(‘CHLexp’,CHLexp)
parameters.put(‘CHLfak’,CHLfak)
parameters.put(‘thresholdRtosaOOS’,thresholdRtosaOOS)
parameters.put(‘thresholdAcReflecOos’,thresholdAcReflecOos)
parameters.put(‘thresholdCloudTDown865’,thresholdCloudTDown865)
parameters.put(‘outputAsRrs’,outputAsRrs)
parameters.put(‘deriveRwFromPathAndTransmittance’,deriveRwFromPathAndTransmittance)
parameters.put(‘useEcmwfAuxData’,useEcmwfAuxData)
parameters.put(‘outputRtoa’,outputRtoa)
parameters.put(‘outputRtosaGc’,outputRtosaGc)
parameters.put(‘outputRtosaGcAann’,outputRtosaGcAann)
parameters.put(‘outputRpath’,outputRpath)
parameters.put(‘outputTdown’,outputTdown)
parameters.put(‘outputTup’,outputTup)
parameters.put(‘outputAcReflectance’,outputAcReflectance)
parameters.put(‘outputRhown’,outputRhown)
parameters.put(‘outputOos’,outputOos)
parameters.put(‘outputKd’,outputKd)
parameters.put(‘outputUncertainties’,outputUncertainties)
result = GPF.createProduct(‘c2rcc.olci’, parameters, OLI)
product=ProductIO.writeProduct(result,‘F:/hydromerit/Hydromedit/images/sentinel_2/out/20180721_S3_C2RCC.dim’,‘BEAM-DIMAP’)
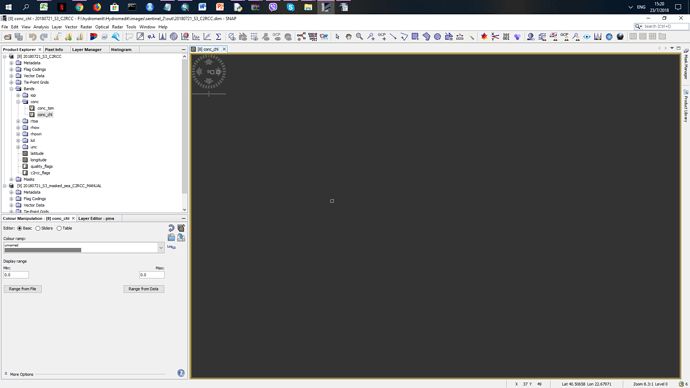
But my output in some bands like concetrations of chlorophyll is Nan (attached image)
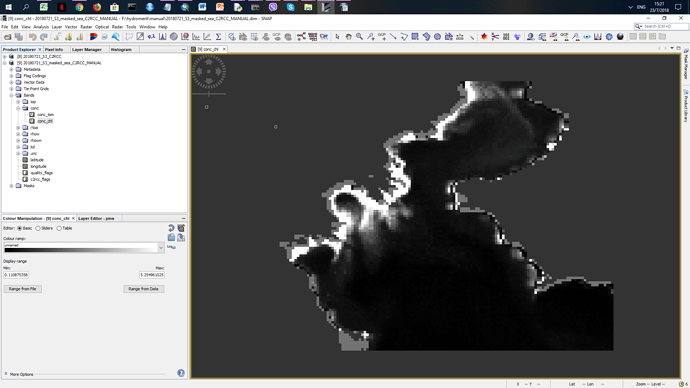
Doing the same preocedure manualy but in the same band i have an output (attaching image):
Any ideas?