Please, explain me more! 
Hi @gabrielaquintana77,
You changed two scripts with new ones. Good. Now, you need to do mt_prep step again.
Fikret
thanks for your reply @Fikretjfm
let me see if I understand, you talks about of make the processing with the command
mt_prep_gamma 20160208 /sdd/RESULTADOS1 0.4 2 2 50 200 again?!
if this is case, I did, but the pscands continued empty!!!
Yes but you changed the name of script to mt_prep_gamma or did you just copied and pasted new script into matlab folder. Because your command should be start mt_prep_gamma_snap not just mt_prep_gamma if you did not replaced with new one.
ah wow!!! let me make again the copy and pasted of the scripts and the run again!!!
Hi @Fikretjfm
Yesterday, I did ran the script again, this time
./mt_prep_gamma_snap 20160208 /sdd/RESULTADOS1 0.4 2 2 50 200
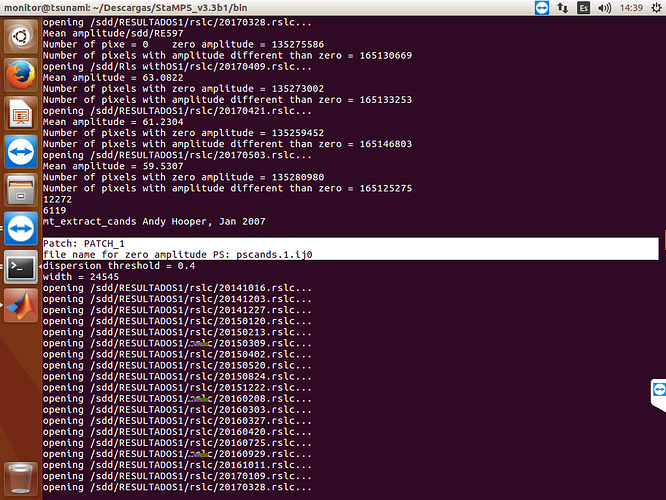
and this time, I can see that executed pscands.1.ij0
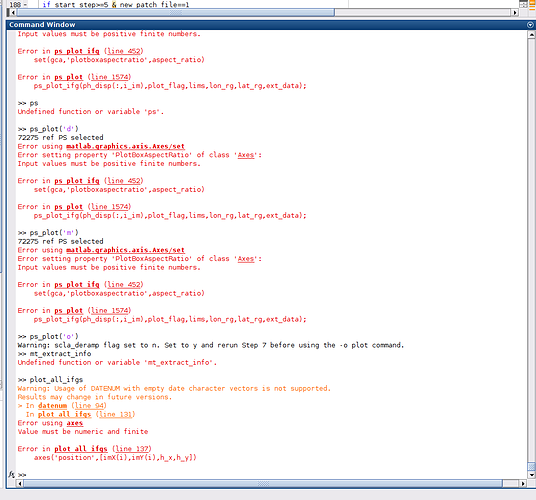
but, I think that the processed not overwrite the PATCH files!!! because this give me error when I want to plot
I readed the suggestion from @thho , so, my question is, I have to modify this script ps_plot.m or getpsver?
http://forum.step.esa.int/t/linux-installation-using-stamps-and-s-1-data/8752/13?u=gabrielaquintana77
thanks very much for advance
Hi,
You need to delete Break commands in some .m files. I will send you my runnable matlab folder. But ı am not at home at this weekend. I will send next Monday.
Fikret
Hi,
We returned back today and here is my matlab folder. Just copy and paste all files.
Fikretmatlab_folder.rar (148.6 KB)
Hi @Fikretjfm
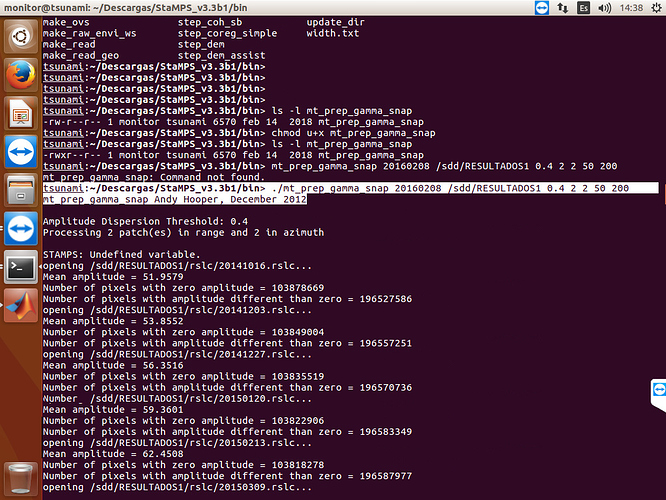
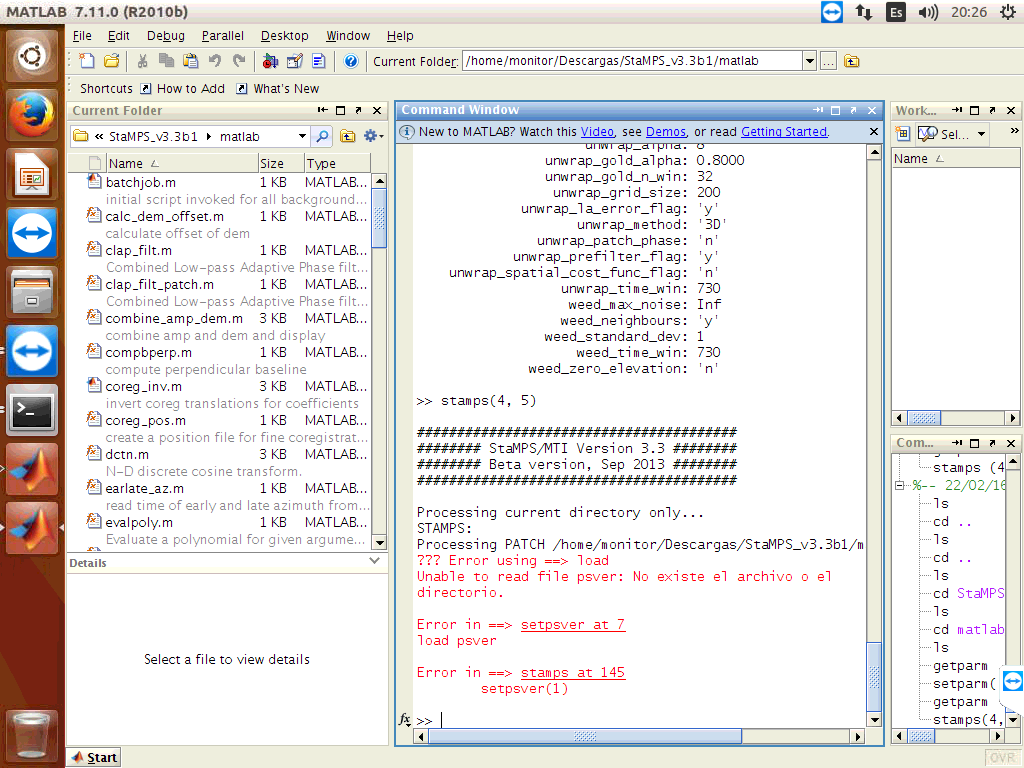
I did run again, with the folder that you send me, and have the same error, the pscands file are empty
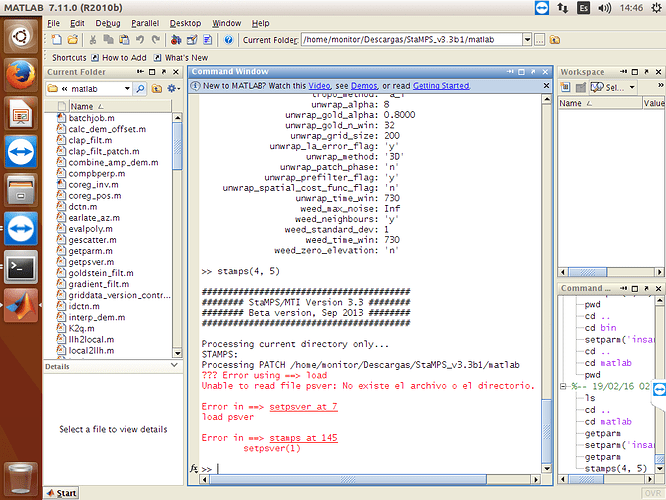
and I obtained error with setpsver!!!
Please I need help with this
Thanks for advance
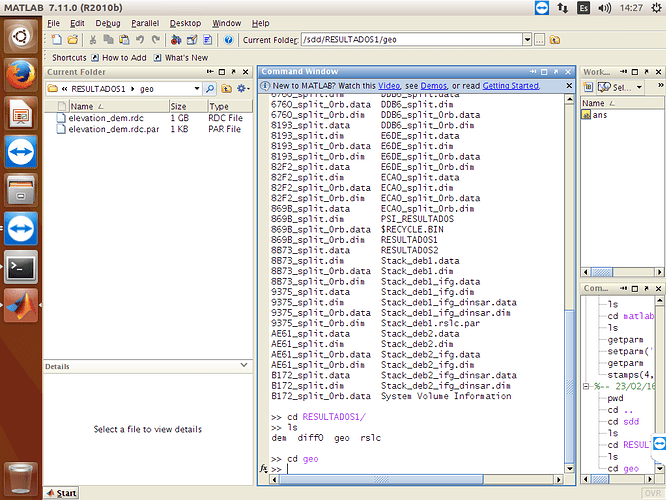
If your .ij files are still empty, it is about running mt_prep_gamma_snap step with wrong script. I am adding you my mt_prep_gamma_snap script try it again. You need to handle with .ij files error first. Do not try stamps step without getting .ij files. And it is related to master.lat and master.lon files in your geo folder. Could you show me your geo folder with files sizes? First show me folder and if it is ok, try following step.
Also I added my mt_… script that works fine for me. Just you need to change at the and of the script with starting grep:
grep -rl ‘/cygdrive/?’ *.txt | xargs sed -i ‘s+/cygdrive/f+F:+g’
grep -rl ‘/cygdrive/?’ *.out | xargs sed -i ‘s+/cygdrive/f+F:+g’
grep -rl ‘/cygdrive/?’ *.in | xargs sed -i ‘s+/cygdrive/f+F:+g’
The question marks is your drive, if you work under /c write c for question marks. Also the second f+F should be c+C. For example like that:
grep -rl ‘/cygdrive/c’ *.txt | xargs sed -i ‘s+/cygdrive/c+C:+g’
grep -rl ‘/cygdrive/c’ *.out | xargs sed -i ‘s+/cygdrive/c+C:+g’
grep -rl ‘/cygdrive/c’ *.in | xargs sed -i ‘s+/cygdrive/c+C:+g’
mt_prep_gamma_snap (6.6 KB)
Hi
We finally find the error. There should be master.lat and master.lon coordinate files. Therefore your .ij files are empty. Export .lat .lon files into geo folder. There are so many explanation exporting them in forum.
Fikret
Ok Fikret,
I am resolving this, when I end this step I will go run again the script 
Thanks for your support
Hi @Fikretjfm
One question, the results that I obtained in the export (/diff0, /geo, /dem, /rslc) they have to be in same folder of results with PACTH 1, PACTH 2, PACTH 3 and PACTH 4???
No not need
Is there an updated workflow for this procedure? I will probably make and share a step by step video if I can replicate good results.
does anyone know how to solve this problem? thanks.
ps_plot(‘v’) ps_plot(‘d’) and ps_plot(’…’) shows error setting property 'PlotBoxAspectRatio ’ of class ‘Axes’ : input values must be positive finite numbers.