the changelog text indicate we should change the work drive,does “the work drive”means matlab install path?
hello~
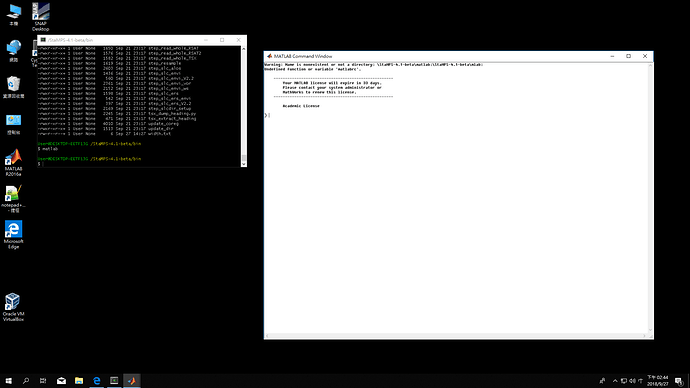
I can use matlab with cygwin64 too.
BUT when I cd to my data catalog then call matlab, it cannot execute…
so I can’t answer your question sor.
you have to navigate to the directory where you exported the files. Based on your screenshot, you are in a different directory. It’s the one conaining PATCH_1 which was generated by mt_prep_gamma.
Hi Bayzidul,
Can we graph process our data upto stamps export?
I have 10 inteferograms generated from Sentinel1 data and want PS InSAR processing.
If you have both master-slave amplitude stack and the master-slave interferogram with the elevation mask and the orthorectified latitude and longitude bands in the output, then yes, you can. Otherwise you are not compliant with the input requirements for the StaMPS export operator.
Otherwise you can try to use the snap2stamps package (https://zenodo.org/record/1308687) with does all the pre-stamps processing for you (only the master splitting has to be done manually using the SNAP GUI). See thread: Snap2stamps package: a free tool to automate the SNAP-StaMPS Workflow
Hello, thank you for your suggestion.
I found a solution of this problem. https://groups.google.com/forum/m/#!topic/mainsar/MDMV9yXDl1g
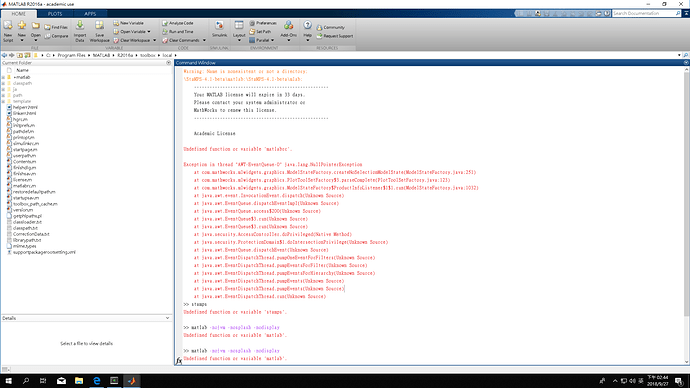
But it’s too simple, I can’t understand how to execute. I have done creat a startup.m and addpath, but I don’t know how to correct “the call of matlab scrip” or my patch01 data path?
Thank you very much
Sincerely
Jamie
Dear Fikretjfm .I met the same problem as your’s.I use the following method and finally sloved it. First, add /staMPS_4.1-beta/bin to the PATH. Second run mt_prep_gamma under the Export folder (where your datas prepared for stamps,may be from SNAP or GAMMA or others).
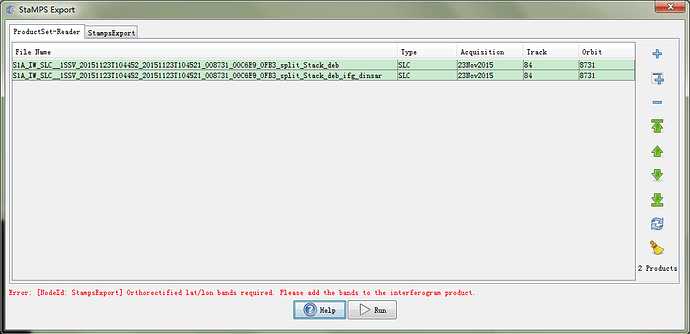
thanks for your reply,a question occurs when StaMPS export, the picture is like this?how do
I do next?
Dear @zhuhaxixiong,
You should re-do the interferogram stacking by adding the orthorectified latitutde and longitude bands on the output. As this information is needed for running StaMPS with the coordinate information. This is possible by using only SNAP v.6.0 onwards. In previous version this is not possible. If you are using old graphs (xml), please update it.
You can get as example the snap2stamps package I have prepared together with M. Foumelis here: https://zenodo.org/record/1322353#.W7r9Iy5uZQI
I hope this helps.
thank you very much
but my interferogram was done in SNAP,the link you give indicates that stamps need DOI?,I didnt install DOI on my computer,and I am not willing to use DOI.
Dear @zhuhaxixiong ,
I met the same question when i use SNAP. Maybe you can try it use my method. Please ensure chack the “Output orthorectified Lat/Lon” option both in interferogram and TopophaseRemoval. Then run it .
I hope this helps.
Lamu
Hi all,
I encountered the following problems when using snap2stamps script. I tried setting the environment variable, but i still haven’t solved it .Maybe i still don’t understand what the ‘current folder’ means. I hope you have time for some guidance.
Thanks
Lamu
have you seen this topic? Snap2stamps package: a free tool to automate the SNAP-StaMPS Workflow
We talked a lot about setting the correct paths in the conf file.
Dear @zhuhaxixiong,
The link I have sent you is a link from which you can download some scripts I have prepared and released on last IGARSS conference which helps on the automatic processing of single master DInSAR using SNAP. In case that you use it and you find it useful, I would appreciate you can reference using that link.
You do not have to install anything on your computer to use it.
Let me know if it is clear now.
Cheers,
Thank you very much!!
I have succeeded step1-7 of StaMPS with Linux, Ubuntu 16.
Thank you for your suggestion
Sincerely
Jamie