delete the whitespace before the brackets:
setparm('insar_processor','gamma');
Matlab (and other scripting languages) are quite merciless regarding such small things.
delete the whitespace before the brackets:
setparm('insar_processor','gamma');
Matlab (and other scripting languages) are quite merciless regarding such small things.
Progressing @ABraun, thanks very much!, was a problem of whitespace…
Now, I have new error, in the script ps_load_initial
Hi @gabrielaquintana77,
Please use the latest StaMPS developer version and not latest releasse from https://github.com/dbekaert/stamps , so in such way you may use already the mt_prep_snap and the latest StaMPS version which guarantees the full compatibility between SNAP output with StaMPS PSI processing.
Prior the latest StaMPS version, when I was using previous ones I have also encountered the same issue than you, and at that time I have created the day_1.in and the master_day.in files if they were not present. Sometimes even the headidng.1.in file as well
In addition, using the snap2stamps scripts from https://github.com/mdelgadoblasco/snap2stamps, makes all the DInSAR processing simpler.
Enjoy your processing!
the page seems to have an error at the moment: https://homepages.see.leeds.ac.uk/~earahoo/stamps/
Do you have another source?
It was already posted! They are now also in Github!
in this case I misunderstood the first sentence because it says
and not latest release from https://github.com/dbekaert/stamps
so I thought this was the wrong one.
ouh! yeah, it could be a bit misleading…
There was a new release, but later, on the developer version that you can download from the github as well, they fixed the ps_load_initial_gamma for the cases in which SNAP saved zero values …
This is needed to avoid further issues with index out of bounds in following steps.
So I wanted to say that both official release from this summer and developer versions can be downloaded from github, but get the clone from the site itself instead of the release .
I hope this is clearer now.
Sorry for the confusion
Hi, dear all, I find that it’s no error if I run command “stamps”, but if I rerun codes like “stamps(5,8)” after the first “stamps” finished, when I draw the displacement map using the codes like the following,
case {‘u2d-dmo’}
uw=load(phuwname);
scla=load(sclaname);
ph_all=uw.ph_uw;
ph_all=uw.ph_uw - repmat(scla.C_ps_uw,1,size(uw.ph_uw,2)) - scla.ph_scla;
subtract of oscialtor drift in case of envisat
ph_all = ph_all-ph_unw_eni_osci;
ph_all =-ph_all*55.5/4/3.141592653;
units='mm';
deramping ifgs
[ph_all] = ps_deramp(ps,ph_all);
ph_all(:,master_ix)=0;
clear uw scla
ref_ps=ps_setref;
fig_name = ‘u2d-dmo’;
there would be error like the following picture:
Thank you !
according to your screenshots no PS are left.
Please check in the previous steps how many were initially selected and left after the dropping of noisy pixels and weeding.
I have checked the ps points left, about 448000. And I run the command “ps_plot” with the parms using “ps_deramp”, like “ps_plot(‘dmo’)” ,the codes are the following,
case {'u-dmo'}
uw=load(phuwname);
scla=load(sclaname);
ph_all=uw.ph_uw;
ph_all=uw.ph_uw - repmat(scla.C_ps_uw,1,size(uw.ph_uw,2)) - scla.ph_scla;
% subtract of oscialtor drift in case of envisat
ph_all = ph_all-ph_unw_eni_osci;
% deramping ifgs
[ph_all] = ps_deramp(ps,ph_all);
ph_all(:,master_ix)=0;
clear uw scla
ref_ps=ps_setref;
fig_name = 'u-dmo';
and set a breakpoint and see how the values change. I found “scla.C_ps_uw” in line 5 is NAN matrix, as a result, “ph_all” had values before line 5 running, and “ph_all” also become NAN matrix after line 5 computed.
One reason could be that the coregistration failed.
ok,thank you
I can’t solve the errors still, what happened to my scla.C_ps_uw…I guess there must be incorrect parms, but it’s a waste of time if I adjust each parm one by one…
Hi all @ABraun @thho @Fikretjfm
Newly I have .1.ij files empty!!! this time I checked my .lon and .lat files, and believe are corrects, I don’t understand the why of my empty files!!! please any suggestions??? thanks for advance
the files inside the geo folder don’t seem correct. The dem files should be in the dem folder and the image data in the rslc folder. Are you sure the StaMPS export was completed successfully?
Have a look here and especially FeiLiu’s comment:
Indeed @ABraun is right. You should get it on different folders (rslc, diff0, dem and geo)
Probably the mt_prep_gamma_snap had not found the proper folder structure.
Had you had a look at the logging of the script itself? Sometimes it might be useful.
Hi
I’m trying to execute STAMPS with SNAP using CygWIN for several days and I didn’t succeeded yet. Tha last problem is that STAMPS in Matlab has error:
Index in position 2 exceeds array bounds. Error in ps_load_initial_gamma (line 108) rg=rgn+ij(:,3)*rps;
I found that the problem is about Patch files. Al pscands*.* files in all Patches are empty!
Is this because if wrong inputs or what?
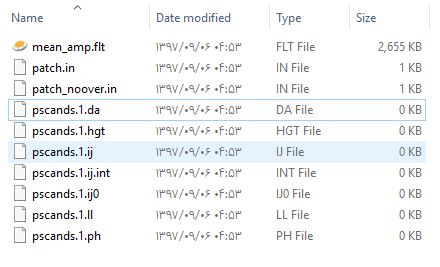
The other weird thing is that “StaMPS_CONFIG.tcsh” is never executed. I changed the path to “triangle” and “snaphu” and nothing changed. I added some error text inside it but no errors occured when running StaMPS. What are these configs?
Should I try to do all the process in Linux?
did you source the config file?
if you work with the tcsh config file, you will have to use the tcsh shell. I also tried a long time to get it done in cygwin but I think that the amount of possible new errors is not worth it. If you have the chance to use Linux for these steps, I highly recommend it. I used a second computer for it or a virtual machine.