Hello,
I’m trying to save Sentinel-3 data as NetCDF format in the ‘Batch Processing (SNAP)’,
but somehow it is saving the file as SEN3 format. Does anyone know why this occurs and how to fix the problem?
Saving the data as NetCDF format works in the ‘Graph Builder’ but not sure why such error occurs in the
batch processing.
For example, in the target Folder below, you can see that I’ve set the “Save as: NetCDF4-CF”.
I’ve also set the file to be save as NetCDF-CD in the graph builder.
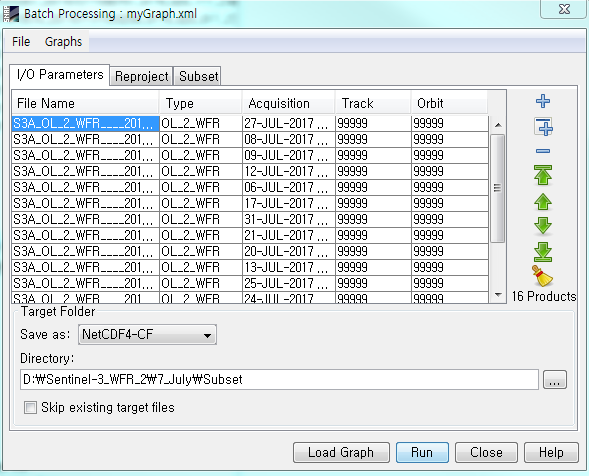
But when I process it, it is saved as SEN3 format:
If anyone knows what I’ve done wrong or how to do fix it, let me know.
Thank you  .
.