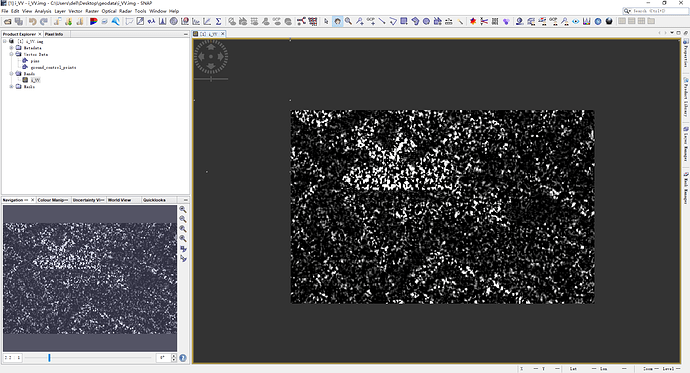
I use SNAP read terrasar .cos file, and create a subet which is named i_VV.img, the result shows like:
then I use Matlab read the same i_VV.img, but there is no object showed, the Matlab code is:
close all;
clear;
clc;
filename = 'i_VV.img';
% data = multibandread(filename, [243, 382, 1], 'int16', 0, 'bsq', 'ieee-le');
fid = fopen(filename);
data = fread(fid, [243, 382], 'int16');
% fclose(fid)
figure; imshow(mat2gray(data));
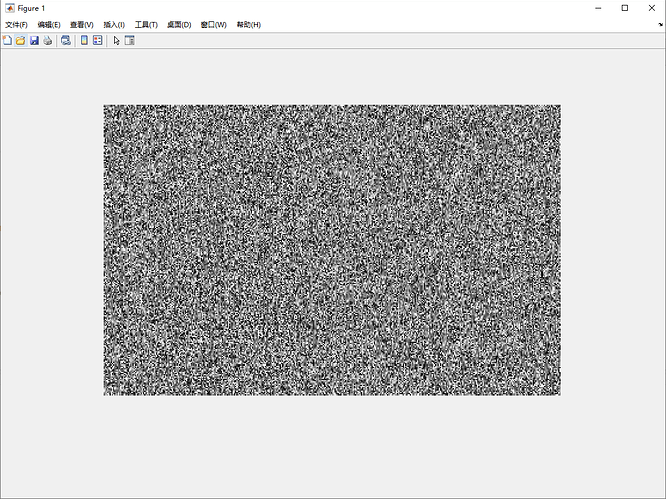
the result of Matlab like:
It seems like the nosie, I try different params, but there are not correct result.
Not only the show img, the values are also different in the pixels. For example,
In SNAP, I select the pixel located in (381,0) > right click > Copy Pixel-Info to Clipboard, the result:
Product: i_VV.img
Image-X: 381 pixel
Image-Y: 0 pixel
BandName Value Unit
i_VV: 46
the value is 46, but in matlab, the data[1, 382] is -15870.
So I want to know how matlab could read the .img file, then letting the value is same to the vaule in SNAP.
Thanks.
The data.zip is here:
data.zip (128.4 KB)