Thank you so much.
Hi @mdelgado Thank you very much for your contribution. I am a beginner and I have a problem, I hope you can help …
When I run slaves_prep.py I get the following error.
File “slaves_prep.py”, line 33
- print P*
Download snap2stamps from the sites you recommended and i am using python 3.8.5
I’m using python2 the snap2stamps code working good.
in the user manual it says that snap2stamps works with python2.7.
in next release it will work with python3.
Dear all,
I have used sanp2stamps with SNAP8.04.
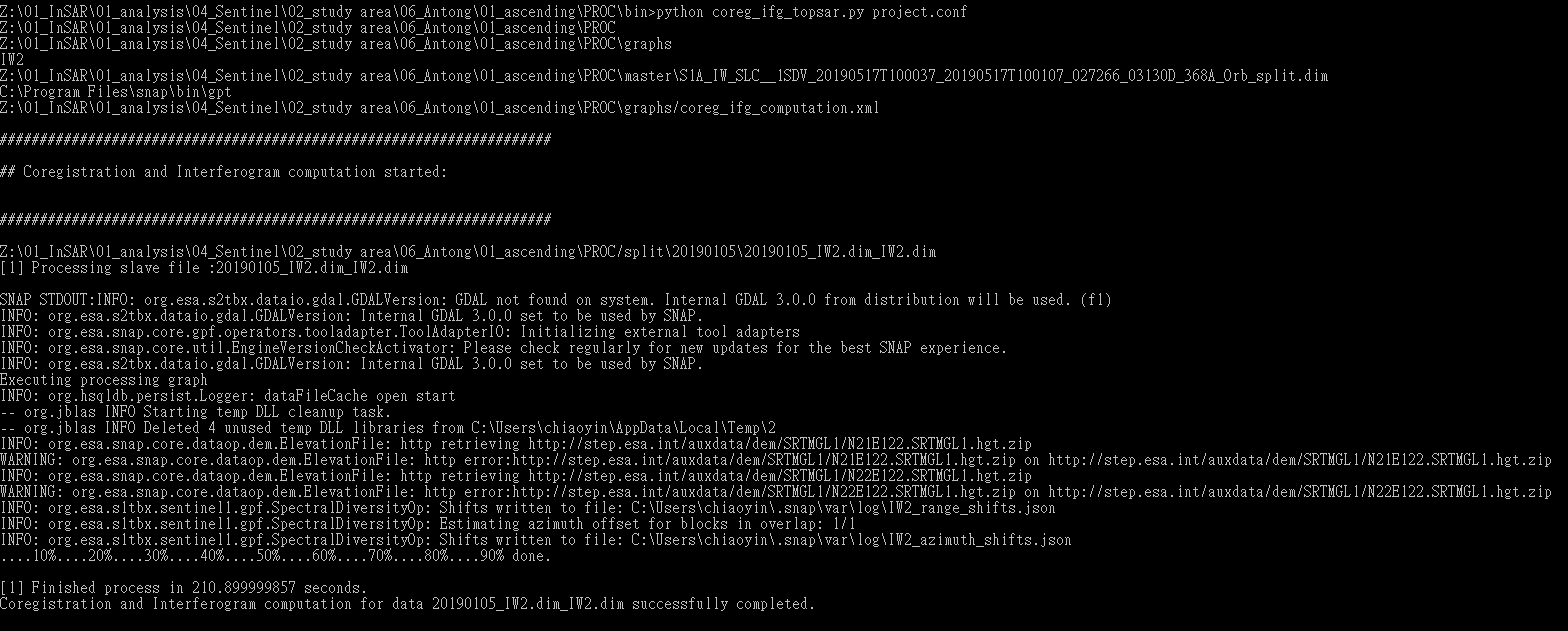
But the slave images in the coreg folder were strange after the step “python coreg_ifg_topsar.py project.conf”. Also, the interferograms were failed.
There were no error message during processing.
The slave image was shown as follow:
Could anyone help me?
Thank you very much!
Hi!
I believe that the master split is same as Master after coreg step. Please check anyway.
What I see on the slave after coreg step… as there is no error in SNAP processing using the snap2stamps scripts, it look to me that it has been any error while writing on disk. Not sure if disk/RAM or JVM -Xmx memory related.
I hope this helps
I’ll check. Thank you very much!
Hi @mdelgado,
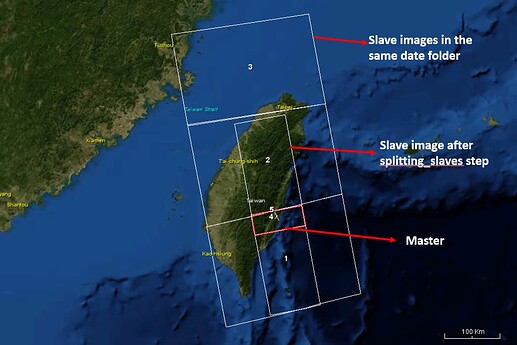
I found out that when there were more than two images in the slave folder on the same date, the burst nearest the boundary of two slaves would be blank after the coreg step without any error message like I mentioned above.
The step “splitting_slaves” will only assembly two images and split it in the folder of three images. But I have checked the split area of the salve covered the area of the master image.
I don’t understand why it would happen. The only way to fix it is keeping only two images in the slave folder?
Thank you for your help.
Sorry but I am not sure I got the point.
Snap2stamps does the slice assembly of maximum 2 slaves contained in same slavedate folder. You can change that manually on the script and xml if needed.
The slice assembly does not write white bursts, that is strange. be sure that there is a spatial continuity with all slaves. Open the original files to have a clear view
If master does not overlap the upper slave image, why you keep it in there? this will need more hdd space, processing time for nothing.
I hope this helps you to overcome your issue.
I have check the original files of all slaves, they are no problem before the coreg step.
Because I have downloaded all images of our country and I didn’t choose the exactly slaves of all date manually, the upper slave image that does not overlap the master area will exist before the coreg step.
Thank you for some opinions for my issue. I will try to overcome them.
Hello!
I am having trouble with the splitting_slaves.py file.
The master image consists of two SLC images, which I processed manually in SNAP, using the graph below:
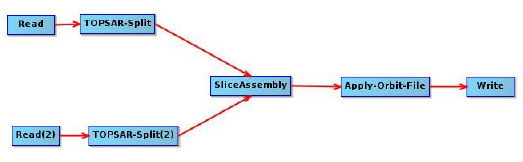
The resulting master has 2 bursts (burst 1 from the 1st image and burst 9 from the 2nd image).
When I run splitting_slaves.py, the resulting slaves are slice assembled correctly, but all the bursts from the two original slaves are included.
How could I make it so that the splitting_slaves.py script returns only the two bursts from the master?
Thanks!!
You can do it by using a tag called wktAoi where you may introduce the BBOX of your AOI in Well-Known Text Format (WKT :e.g. POLYGON (( … )) )
This will be introduced in the next snap2stamps version
Best
Hi @mdelgado! I just now found the time to try this out. It works like a charm!
I also found this post by @andretheronsa who was kind enough to share the modified scripts incorporating the AOI polygon.
Thank you very much!!
I’m not sure maybe this site is of any help:
Wicket - Lightweight Javascript for WKT [Sandbox] (arthur-e.github.io)
MS Excel does the job as well 
Hi,I‘m a user of the SNAP and StaMPS. When I process the different frame Sentinel-1A SLC data coregistration with Python and snap2stamps, I met a problem that can not estimate offset in overlap with the different frame data. Can you help me out of this?
Thank you very much!