Hey guys,
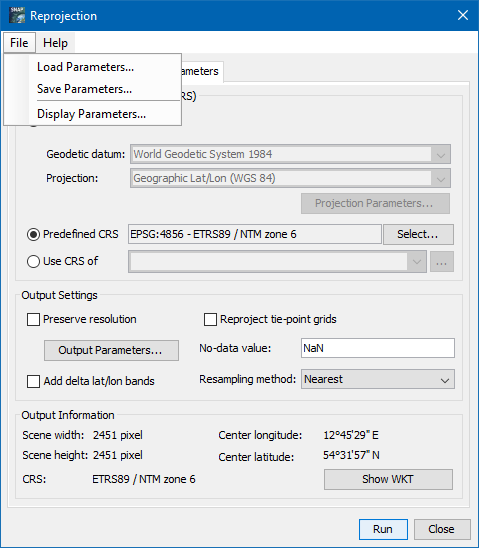
I need to process a large amount of Sentinel-3-Syn-2-data and I’m trying to do it with Snappy. I have some experience with Snappy when it comes to processing Sentinel-1-data and wanted to kind of reuse these functions I wrote back then. To get a clearer picture of what I want to do: I just want to georeference and export (a subset) in netcdf format. In the GUI it is quite easy: 1. Open the ‘xfdumanifest.xml’, 2. Reproject with Raster/Geometric/Reprojection: tick off ‘save as’, predefined CRS, bicubic, preserve resolution, tick off ‘reproject tie-point grids’, 3. select product, export, select spectral bands and flags, create a subset from geo coordinates, export as netcdf. Done.
If I translate that simple workflow to snappy it goes as follows:
1. Read Product
def read_sentinel_product(manifest_file_path):
r = ProductIO.getProductReader('SENTINEL-1')
p = r.readProductNodes(manifest_file_path, None)
return p
Obviously with Sentinel-3 it is another ‘product reader’ but the also obvious solution ‘SENTINEL-3’ does not work. So which reader am I supposed to use?
2. Reproject
The basic skeleton of every snappy-function is:
param = HashMap()
param.put(...)
...
GPF.createProduct(FuncNameAsString, param, InputDataSet)
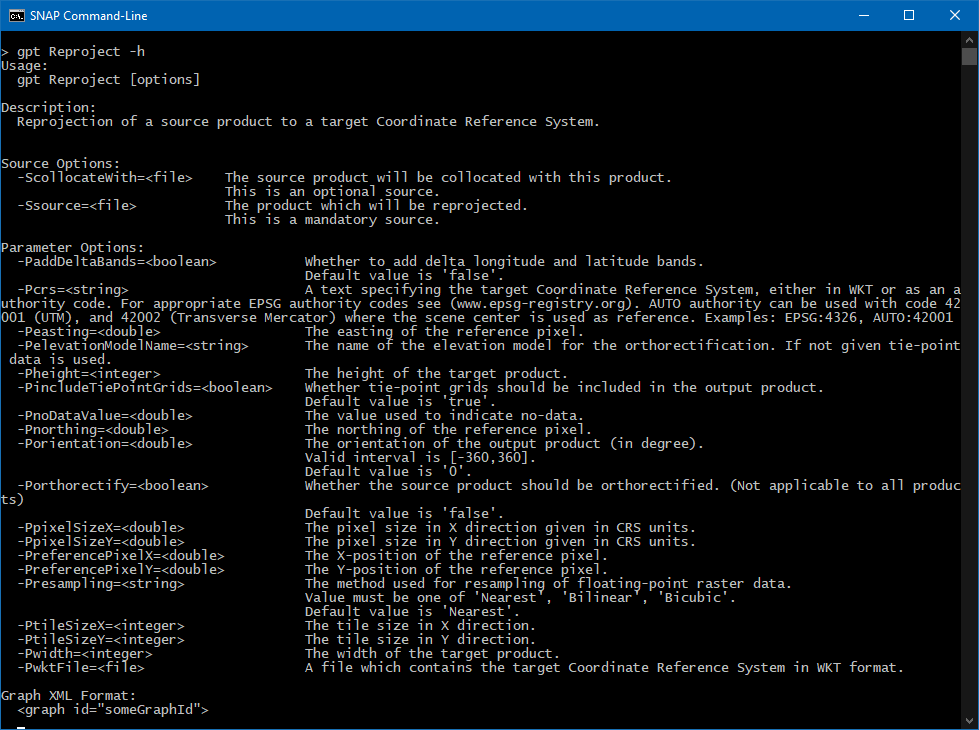
If I iterate over the list of Operators, possible parameters for reprojection are:
wktFile or None
crs or None
resamplingName or resampling
referencePixelX or None
referencePixelY or None
easting or None
northing or None
orientation or None
pixelSizeX or None
pixelSizeY or None
width or None
height or None
tileSizeX or None
tileSizeY or None
orthorectify or None
elevationModelName or None
noDataValue or None
includeTiePointGrids or None
addDeltaBands or None
I couldn’t find anything about how a CRS is correctly passed as parameter. My guess would be:
param.put('crs', 'EPSG:4326') # is that correct?
I wouldn’t say that the rest of the list above is self explanatory, but the (for my use case) relevant settings would be (I guess):
param.put('noDataValue', 'NaN')
param.put('includeTiePointGrids', False)
param.put('resamplingName', 'Bicubic')
3. Subset
My old function takes a wkt-string. I would rather give Lon/Lat coordinates but again I don’t know the correct format. I guess the correct parameter is named ‘geoRegion’. In that case do I pass a list ['Lat_min', 'Lat_max', 'Lon_min', 'Lon_max'] or something else?
def create_subset(input_data, wkt):
param = HashMap()
param.put('geoRegion', wkt) # I'd prefer passing coordinates
param.put('subSamplingX', '1')
param.put('subSamplingY', '1')
param.put('fullSwath', False)
param.put('tiePointGridNames', '')
param.put('copyMetadata', True)
subset = GPF.createProduct('Subset', param, input_data)
print('Subset created.')
return subset
Furthermore I would like to add bandNames as I don’t want the whole data set, just the spectral bands and flags. In the GUI I simply tick or untick the bands. How can I select specific bands?
4. Write Product
The last step is writing the product to disk. Atleast here I don’t have any questions, I just added this for the sake of completeness.
def write_product(sat_data, save_dir, save_name, file_format='NetCDF4-CF'):
print('Writing product...')
target = os.path.join(save_dir, save_name)
ProductIO.writeProduct(sat_data, target, file_format)
print('Product written!')
Thanks for your help in advance, I hope somebody can help me with that.
Cheers