This is because it is not possible to choose which reader will be used after the product is computed and opened is SNAP automatically, but the product should be ok. You can try to re-open it in SNAP and choose the appropriate reader (60m if the product was computed at 60m, etc.). Can you confirm the product is ok if you re-open it?
Re-opening fails, regardless of which reader (10m, 20m, 60m).
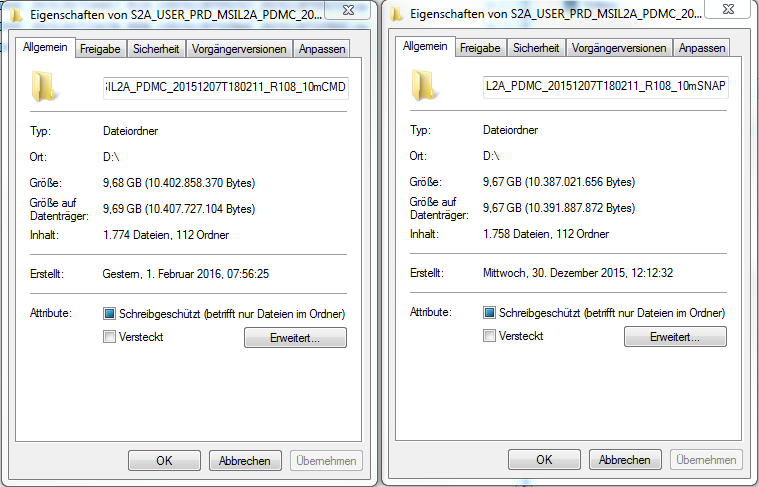
Comparing the L2A datasets, the one from the command-line processing is a little bigger and contains more files.
Additionally, in the xml-file in the root folder entries of S2A_USER_DEM_…60m in ‘General_Info - L2A_Product_Info - L2A_Product_Organisation - Granule_List - Granules’ are missing. The S2A_USER_DEM_L2A_…60m.jp2 files in the S2A_USER_PRD_MSIL2A_PDMC_20151207T180211_R108_10mCMD\GRANULE\Tiles\AUX_DATA folders have only been produced in the command-line processing. In both cases, the folder for DEM storage in the L2A_GIPP.xml has been set.
Regards,
Ralf
SNAP is just launching the SEN2COR from the command line and the SEN2COR is generating the output files so it shouldn’t be different.
What option do you use to launch SEN2COR from the command line?
L2A_Process --resolution 10 D:…(folder)
Hi!
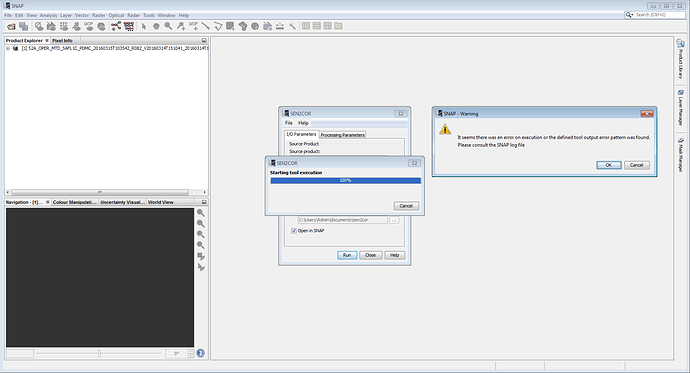
I’m trying to run SEN2COR on Windows 8, but the run starts running the process but not after it ends and the window closes without giving any result
What may be due this?
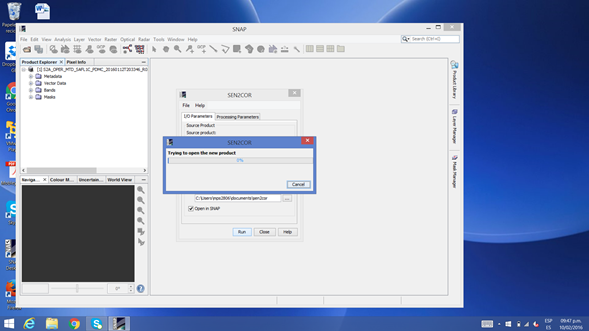
Thank you
Marcela
What resolution did you use to perform the correction?
For the 10m resolution the algorithm has some problem and you can easly found workaround in this forum.
I followed all the installation guides and I was able to run everything in a desktop with win 10.
Are ale the environment variable correctly set?
and do you use the Anaconda python distribution?
i have the same problem as you how can i resolve it pleaaaaaaase
(Meta: I suggest we close this thread to force people to open individual threads for their questions – there is a lot of wild growth here)
i had the same problem i’m using windows 10 and sen2cor 2.0.6 how can i resolve this problem ??
I had the same issue as well, no longer use the Sen2core plug-in, now use the command prompt or windows PowerShell, PowerShell works better, all you have to do is type in L2A_Process --resolution “either 10,20 or 60” folderlocation. Please do not use the quotations, also for 10m fix you have to update the L2A tables as shown in 1 of ChristianSeverin’s updates.
I have the same problem what can i do please !!
hi I’m trying to run SEN2COR but i am having same problem …if you have any idea about this please i need help 
I received that error in cmd (Windows):
C:\Users\user>L2A_Process
Traceback
(most recent call last):
File “E:\Anaconda2\Scripts\L2A_Process-script.py”, line 9, in
load_entry_point(‘sen2cor==2.0.6’,
‘console_scripts’, ‘L2A_Process’)()
File “E:\Anaconda2\lib\site-packages\setuptools-20.3-py2.7.egg\pkg_resources__init__.py”, line 549, in load_entry_point
File “E:\Anaconda2\lib\site-packages\setuptools-20.3-py2.7.egg\pkg_resources__init__.py”, line 2542, in load_entry_point
File “E:\Anaconda2\lib\site-packages\setuptools-20.3-py2.7.egg\pkg_resources_init_.py”, line 2202, in load
File “E:\Anaconda2\lib\site-packages\setuptools-20.3-py2.7.egg\pkg_resources_init_.py”, line 2208, in esolve
File “E:\Anaconda2\lib\site-packages\sen2cor-2.0.6-py2.7.egg\sen2cor\L2A_Process.py”, line 5, in
from tables import *
File “E:\Anaconda2\lib\site-packages\tables_init_.py”, line 84, in
from tables.utilsextension import (
File “__init__.pxd”, line 155, in init tables.utilsextension (tables\utilsextension.c:17736)
ValueError: numpy.dtype has the wrong size, try recompiling
I think that it’s a problem with version compatibility. It was tested on two
computers with Windows 7 and Windows 8.1 with different versions of anaconda
and python. On the one of these computers it was working earlier but I decided
to reinstall. The second was an almost clean system.
Can anybody help me? I’m not sure how (and what) to recompile, but I think
that it is a deeper problem.
Can you select BigTIFF instead of TIFF as output? The maximum file size of 4GB was exceeded in your example.
I was having the former pathing error, but upon correcting it I seem to have found a new one:
Has anyone experienced this or found a solution? I am not sure how to find the log file or what to look for when I find it.
Long path error!
“Long path tool” is very helpful for this problem. You can solve this
problem like copy, delete, long path files by using this tool.
I used to have similar problems too, but after using “long path tool”
everything was solved.
Hi, I have the same problem,unfortunately I haven’t solved the problem yet.Please tell me if you can solve this problem.Thanks.
Install problem (there seems to be a lot of those): I’m trying an install on a Windows machine (x64, Windows Server 2012 R2). I get an ‘ImportError: No module named glymur’. When I check the site-packages of my Anaconda2 installation, there is indeed no module named glymur. And when I try installing it manually (conda install glymur) I am told that there is currently no glymur module in Anaconda’s x64 channel. Please help. I have SNAP and sen2cor successfully installed on another Windows 7 machine, so I know if can be made to work.
Anders