Dear Jaroslav and juanesburgo,
I managed to fulfill export to StaMPS when I re-run again Interferogram formation and TopoPhaseRemoval for the whole co-registered stack. It is not long.
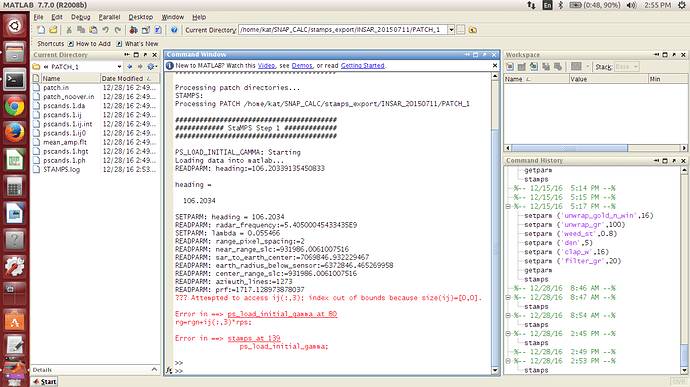
But I still have the problem with the exported data. Below are the values which are imported by StaMPS. Heading is definitely wrong. Moreover look angles are not imported… Could you, please, send me your screen of Step 1 of StaMPS? Or probably you can give me any advise.
Thank you for your time
Katherine
jaroslav
December 28
juanesburgo:Using SNAP. It’s the safest way, I think!
Then SNAP already renames all i and q bands itself
juanesburgo:
Do you have a reason to choose 16 or it was by chance?
I had this error message during Step 6 :
Minimum dimension of the resampled grid (17 pixels) is less than prefilter window size (32). So according to one mainsar forum topic I have changed the value to 16.
Visit Topic or reply to this email to respond.
In Reply To
juanesburgo
December 28
Using SNAP. It’s the safest way, I think!
Like me. I also had to change that parameter. I never studied that step. Do you have a reason to choose 16 or it was by chance?
Visit Topic or reply to this email to respond.
To unsubscribe from these emails, click here .
All the best,
Katherine