Hello @jimmy,
Do you have this message with any S2 L2A product or a specific one?
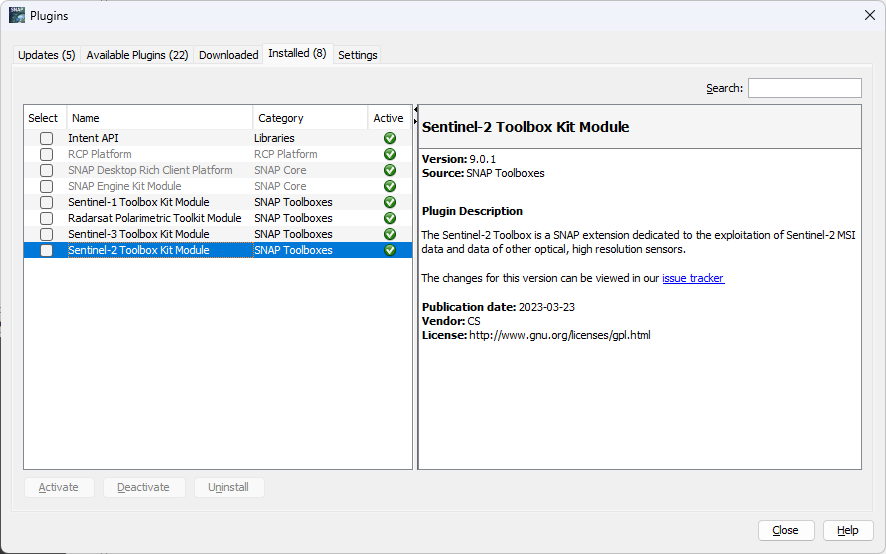
Can you check if “Sentinel-2 Toolbox Kit Module” is the list of installed plugins (Tools → Plugins → “Installed” tab)?
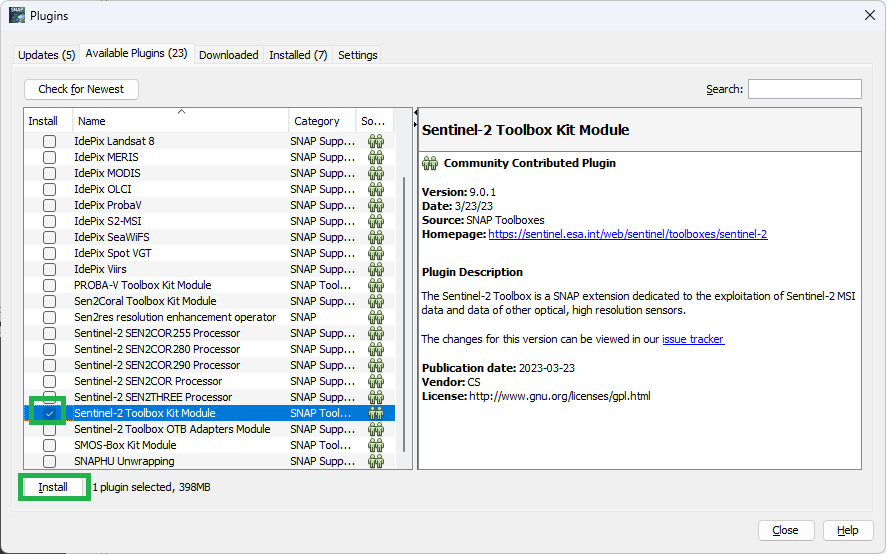
If it is not, it can be installed from the “Available Plugins” tab: