Hello,
I m using S2-L2A product.
I m interested on vegetation and bare soil only, and I d mask (set to NaN) the other classes.
How can I do that with all the bands together. With Band Math, I have to process one band a time.
Also, for filtering, I would filter all the band in the same time.
Thanks
That’s the way you have to do it. Use Band Math on each band. You can also create a graph XML file and use it on the command line.
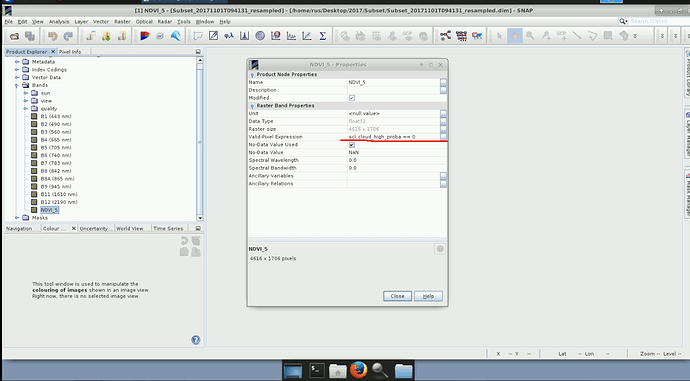
You could also change the valid-pixel expression for each of the bands (select properties from the context menu of a band).
On the command line the Image-Filter operator could be used.
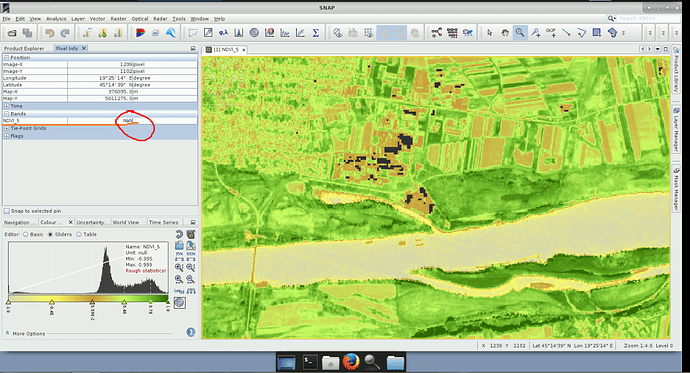
gpt Image-Filter -hIs it possible to calculate NDVI for L2A product using Band Math and on newly created NDVI band use Valid-Pixel Expression, for example, sci_high_cloud_proba == 0 (Pic.1) where High cloud probability pixels are given NaN values for NDVI (Pic.2)
Thank You in advance
What does it do? Should that be clear all the cloud on the map?We need a clear viewon the map so that we can compute NDWI and FAPAR.
Im using SNAP and I want to remove all the clouds for a better result of the map image to compute fapar and ndwi,but hiw can I get rid on the clouds?
Have a look at this thread:
Dear Stephanie, could you explain in more detail your solution. I have the same problem as you. I first created mask by union of several of the build-in masks. Then used that mask in the “Masks->Land/Sea Mask” tool to apply my mask to all Sentinel spectral bands at once (resampled in advance to common resolution) and saved the output as DIMAP. But the resulting images are all black with here and there values 1 or 0. How did you manage to resolve this problem? Many thanks in advance for your help.
Dear Petar,
I went back to my notes and below is what I did.
- Open the Sentinel level 2 product in SNAP (I used the version 5).
- Resample the product
Raster/subset, click OK in the dialog box
In the resampling parameter tab select the 1st option “by reference band” and select B1
In the 1/0 tab: select “open in SNAP” only - Create the mask based on band B1 (this was the one with higher contrast in my case)
Use Mask manager with [x] based on value range. You will need to play around this one to find the value that works for you. - Create a virtual band based on the mask
Go to the mask tab in the product and right click on the one you just created
Select “Band Maths” while keeping the default expression as is which will lead to the pixels under the cloud given a value of 255 while the rest will be set to NaN - Apply the mask on the resampled product and export as GeoTIFF+xml
Raster/Masks/Land/sea mask
In the resampling parameter tab select"virtual band," and use “Vector as mask”, select your cloud mask
In the 1/0 tab: select “save as GeoTIFF+xml”
Hopefully that should do the trick for you as well!
Best regards,
Stephanie
Hi
I’m having problems with the tool Idepix Sentinel2 -MSI tool.
I’m processing a large number of images. All of them were processed in a batch with the following algorithms:
- Subset to band2 pixel size
- subset to the same geographical coordinates.
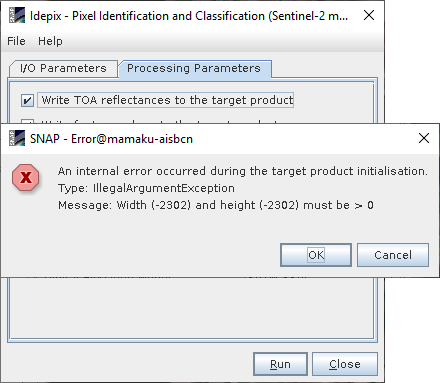
After this, if I apply Idepix tool gives me the following error:
Message: with(-2102) and height(-2102) must be > 0
The weird thing is that this message appear in some images, not in all the processed images. What could be the problem?
Thank!!
are your image L1C or L2A,?
Did you apply Idepix for each single one? Or Did you use batch?
Because not all your images have the error,
this could be related to the UTM coordinate system.
Try reprojecting one raster to WGS84 (EPSG4326) and see if the error persists.
I’ll show you the process:
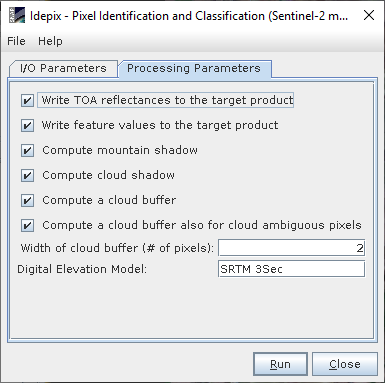
I suspect about the Coordinate system, the weird thing is that this error appear in some images and not in other, when all the images where processed with the same batch process with the same parameters.
area all images from the same tile?
Because some tiles range over two UTM zones, thus some negative coordinates can occur.
No, not all the images are from the same tile.
Could you apply idepix individually to a couple of your image, these have error
Actually, the error happens because of the sun zenith.
@marpet
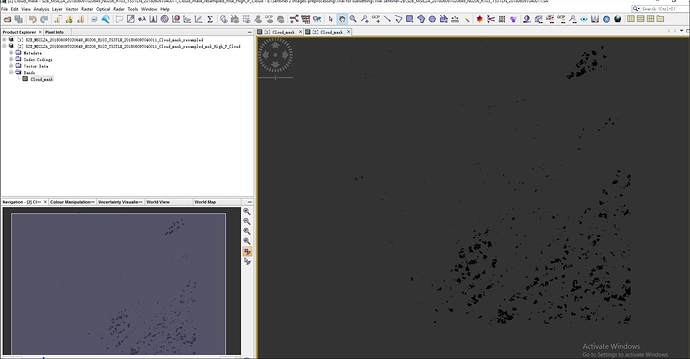
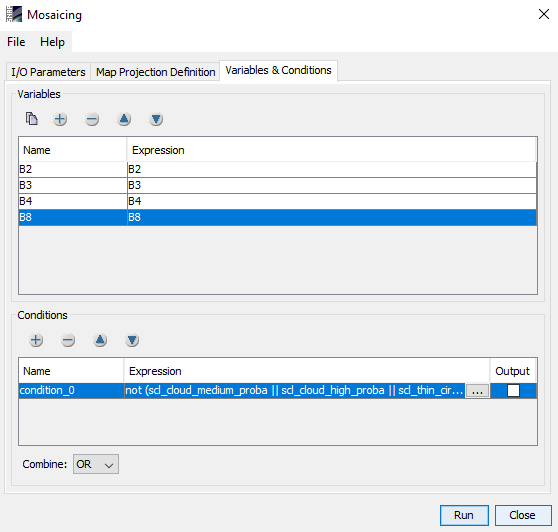
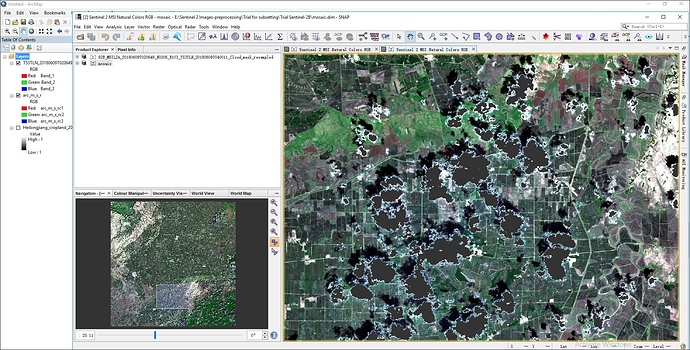
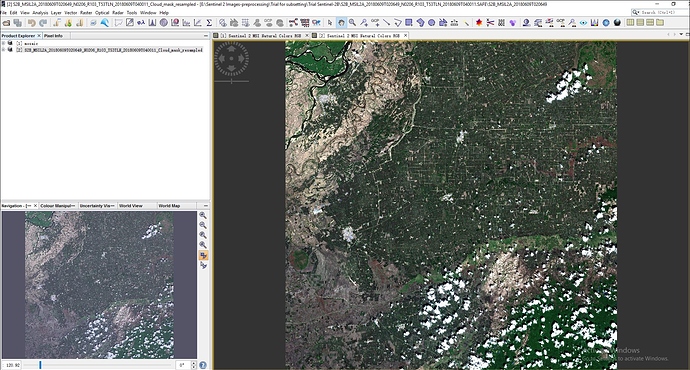
i have done cloud mask for sentinel 2B L2A image in SNAP(Sen2Cor) by creating cloud mask for 10 m resolution using Band Maths for scl_cloud_high_proba, scl_cloud_medium_proba and scl_thin_cirrus separately. Now i want to replace the cloud mask areas by using another clear image as reference image. Could you tell me the process or steps to replace the cloud in SNAP- 6 version. thank you in advanced. i upload here only two separate cloud mask images
If both, the cloudy and the clear reference scene, have the same extent and at the same location (same S2 tile) then you can also use Band Maths.
Use as expression the following:
IF (scl_cloud_high_proba || scl_cloud_medium_proba || scl_thin_cirrus) THEN $2.B01 ELSE B01
If they don’t have the same extent then you would need to collocate them first. and then apply the expression.
But you can also do a mosaic.
@marpet, Thank you so much for you help. I have done Mosaicing of cloudy sentinel 2B_L2A image. i have attached the cloud masking image here. please tell me how can i replace the mask areas with reference image. Thank you in advance.
if both the masked and the reference image have the same pixel extent and size, you can replace the masked values in the band maths with an IF statement as demonstrated here: Replacing NaN with 3x3 average