Dear Katherine,
I keep getting the same error in both Windows and Linux and I think my data structure is somehow not correct:
What I did:
- create S1 stack with lat/lon bands (later placed in the geo folder)
- create interferogram, removed topographic phase, and added an elevation band
- StaMPS export creating dem, diff0, geo, rslc
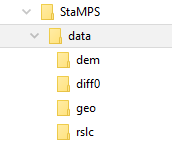
dem folder
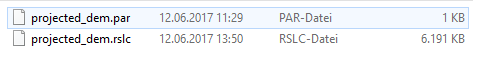
diff0 folder
geo folder (here I am not sure if i named the lat/lon files correctly)
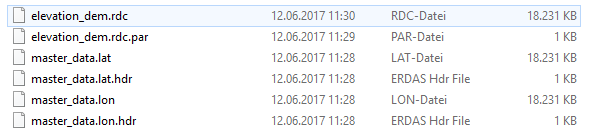
rslc folder
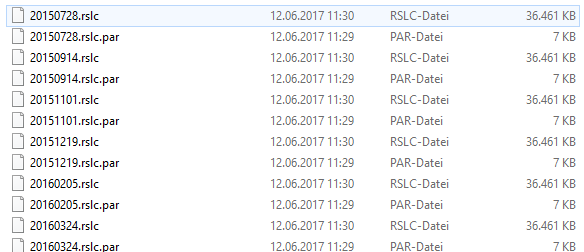
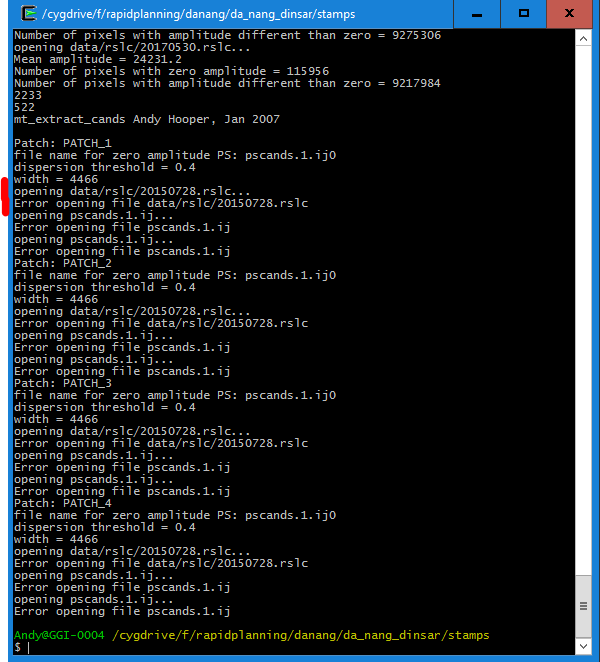
I then open the linux terminal (or cygwin respectively) and run
mt_prep_gamma 20150728 data 0.4 2 2 50 200
This works fine until the selection of persistent scatterers (mt_extract_cands)
It is stated that data/rslc/20150728.rslc cannot be opened but it is located in that directory and was also correctly used in the previous step (mt_prep_gamma).
Am I missing something? Did i run the script from the correct location or should the file structure somehow different? I also looked in the code of mt_extract_cands but couldn’t find why the file cannot be opened.
The same happens for mt_prep_gamma_snap which you provided in your post.
Any hint would be helpful.