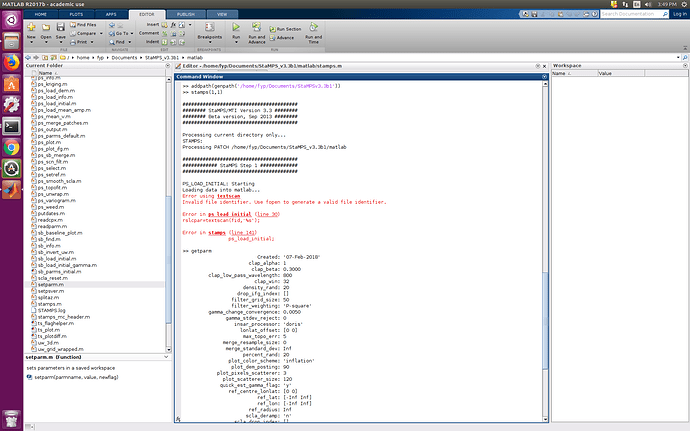
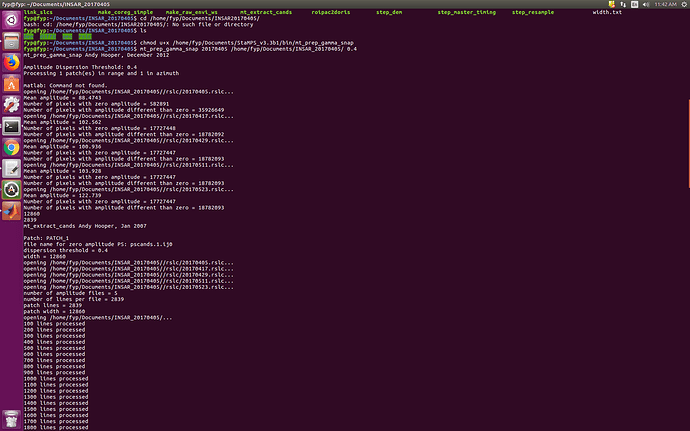
I also have the same results after @thho has helped me with the successful installation.
I have renamed my ps_initial_…(changed) file to the ps_initial file and removed the older “initial” file.
I have also done what some may have suggested as change the gamma in the getparm/setparm.
before I have made these changes, my error looks like this: PSI: Error on first step on Matlab
@ABraun I am using Linux Ubuntu but I also received the same error as you. Any new findings?
I am looking to view PS in highly urban area but there are a few nature reserves around, hence there should be obvious non PS areas in my project if it does succeed.
If anyone is stuck at the Linux phase of the installation, you can view the thread here Linux Installation using StaMPS and S-1 data