Hello jcoca,
I have installed ICOR S2 in my windows pc and i tried to run it…i got the following error…
parsing arguments
done
Traceback (most recent call last):
File “c:\Program Files\VITO\iCOR\src\icor.py”, line 108, in
raise Exception(“environment variable ICOR_DIR not set”)
Exception: environment variable ICOR_DIR not set
Process exited with value 1
Finished tool execution in 0 seconds
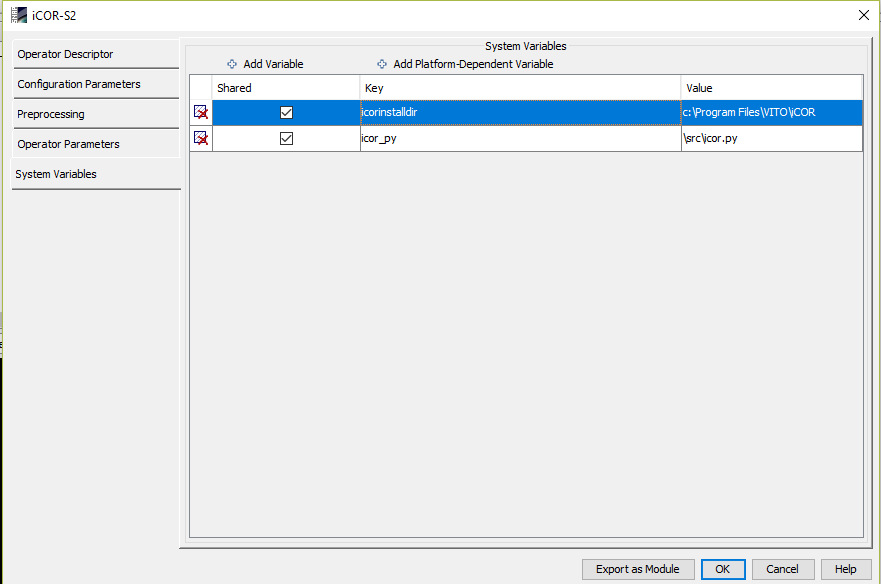
and my system variables are
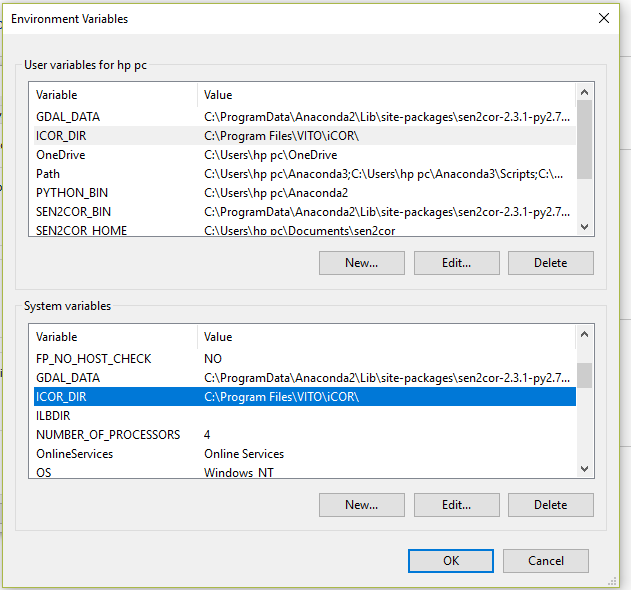
environment variables:
Please help me out to solve this issue…
Thanks in advance…