I’ve investigated this issue and it some that two things come together here.
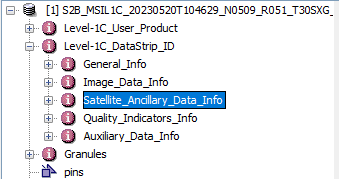
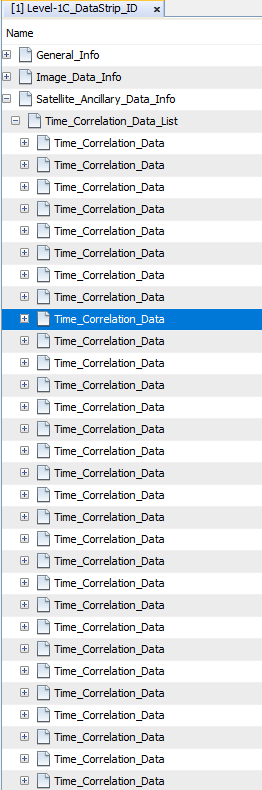
NetCDF-CF has a limit of 32768 groups and S2 is structured in a way that for some products this number is exceeded. We will further work on finding a solution. It might be, that we need to restructure the metadata of Sentinel-2. Especially the data which is currently shown under Satellite_Ancillary_Data_Info:
But this is further under investigation.
For the time being, if the metadata is not important for you, you can remove them by adding a subset operator to the graph.
<node id="removeMetadata">
<operator>Subset</operator>
<sources>
<source>input</source>
</sources>
<parameters>
<copyMetadata>false</copyMetadata>
</parameters>
</node>
In SNAP Desktop the metadata node Satellite_Ancillary_Data_Info can simply be removed by pressing the DEL-key.