How to check it, i’ve never used linux before
Don’t take this personal, this rather concerns all users in tje StaMPS category, but if you want to use programs and scripts bases ob linux you should bring the motivation to google such smaller things by yourself. This forum is not a computer course 
Hi aziz10,
Firstly, I have also never used linux before I have embarked on this project so I think it does help to google stuff online and trying to reference some “linux for dummies” tutorials. I also did not screenshot my ‘gawk’ installation process so I cannot help you to monitor step-by-step. But now, I have managed to learn to download ‘gawk’ on my own with googling so I think you can too 
Nevertheless, you can consider this http://web.mit.edu/gnu/doc/html/gawk_18.html
If it helps, i think you should do this tutorial provided by @thho step by step file:///C:/Users/User/Desktop/snap_stamps_1_0.html#27_matlab
do not miss out any steps because you will get frustrated with the errors if you do. (I personally made these mistakes by myself so I know)
Goodluck!
Chong
Thank you, i’ll try
Can you share your matlab directory or ps_plot.m please?
ps_plot.m (56.1 KB)
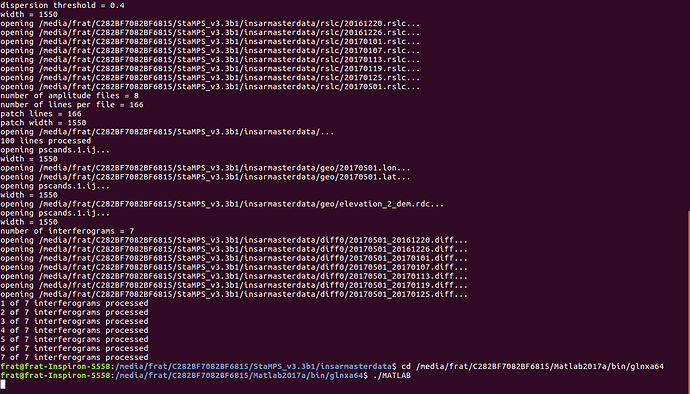
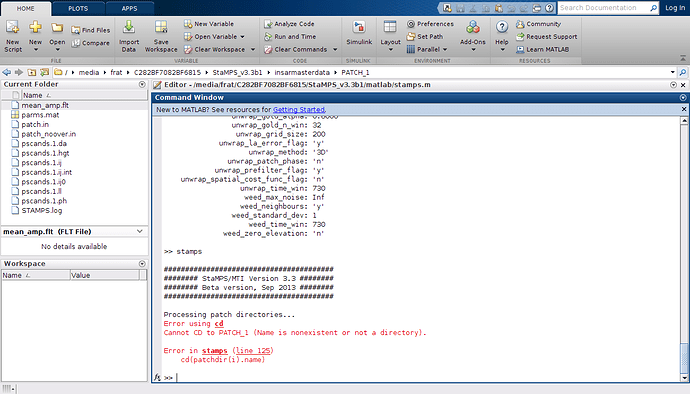
Hello Dear Helpers, ı am trying to do PSI application on my S-1 images and ı am using linux machine and ı can not pass a step can you help me ? @thho @hchong005 @ABraun @FeiLiu @bayzidul @Fikretjfm
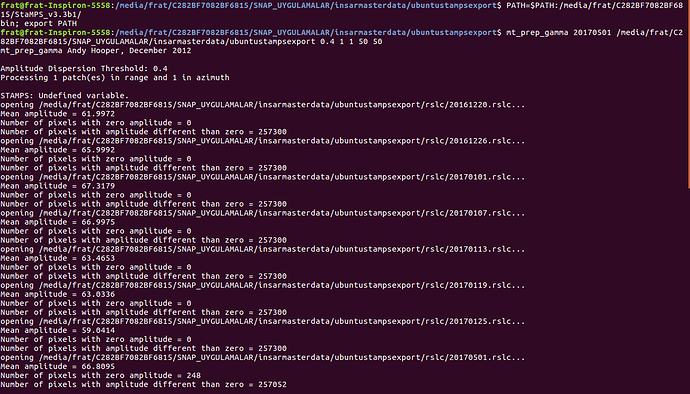
after working terminal just like below
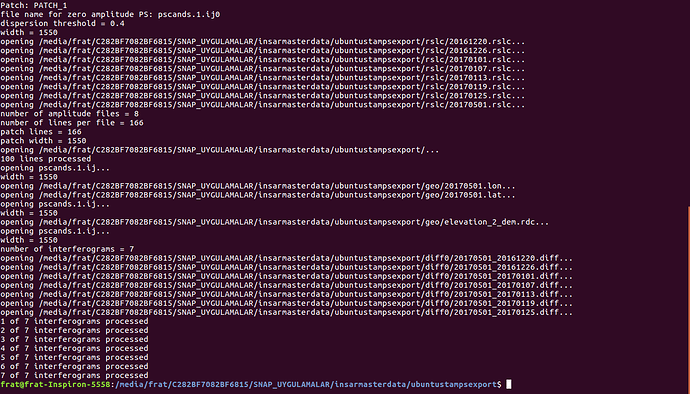
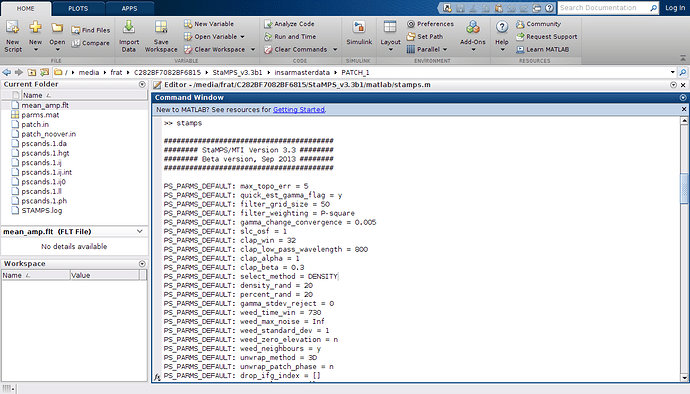
ı am opening matlab and then try to work matlab but it doesn’t work like below
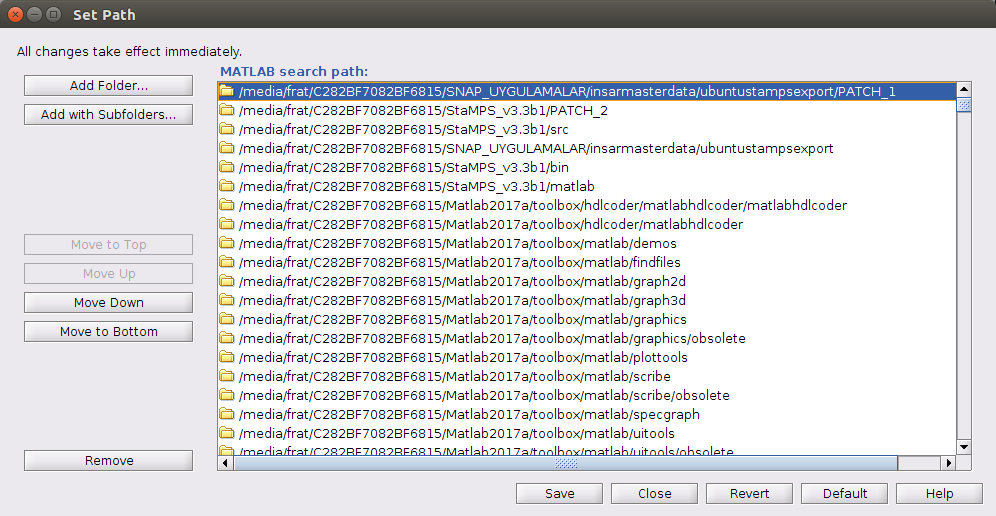
and ı added the my export and Patch datas directories to matlab like below too
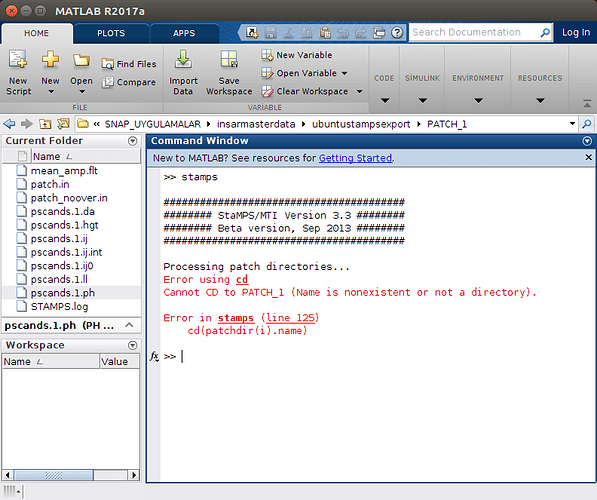
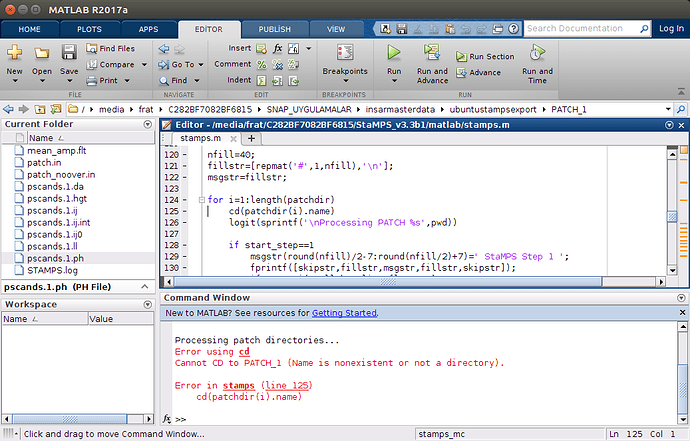
and when ı look at the error, it shows me the picture like below
So, what should ı do ?
Dear @firat,
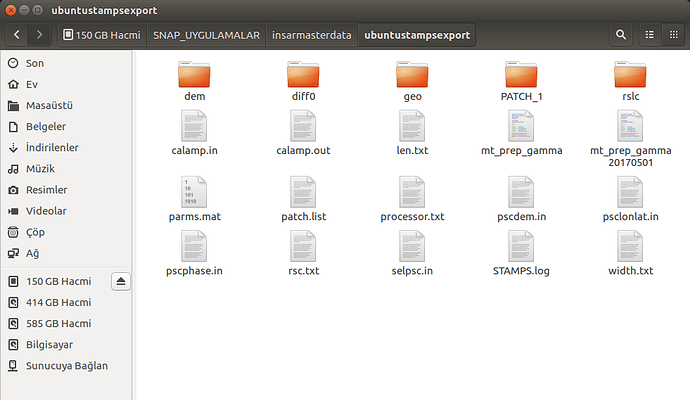
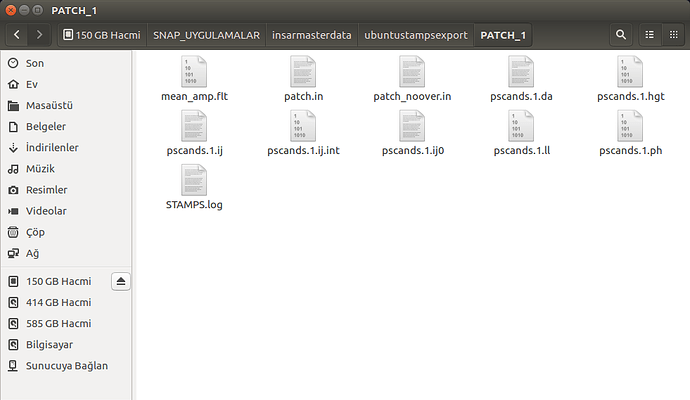
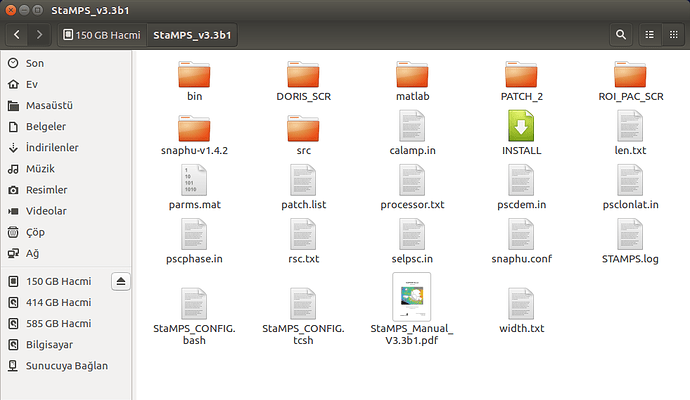
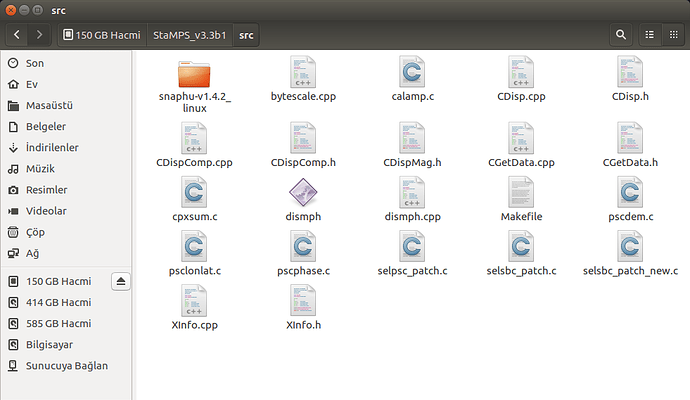
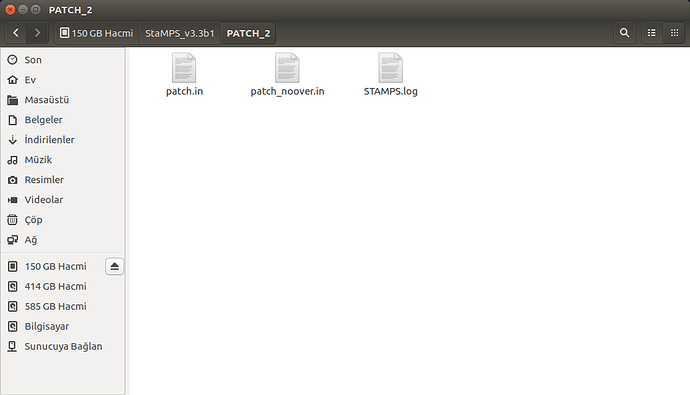
please show us your folder structure of …/insarmasterdate/ubuntuexport/ and …/StaMPS_v3.3b1. I am not sure if everything is ok here because there seems to be exported data (a PATCH_2 and a src folder) in your StaMPS_v3.3b1 directory. A screenshot of your StaMPS_CONFIG.bash might help, too, but since you were able to call mt_prep_gamma_snap and matlab I think this will be ok.
What I do when I have problems with the first steps, I recommend to delete the whole export INSAR_master_date directory, go back to SNAP open stack and ifg again, make a new StaMPS export, and try again starting with mt_prep_gamma_snap etc. For some reason, this fixed my problems the most of the time.
Good luck!
Hello Dear @thho,
Thank you so much for the advise and answer
These are my /insarmasterdate/ubuntuexport/ and …/StaMPS_v3.3b1 folders
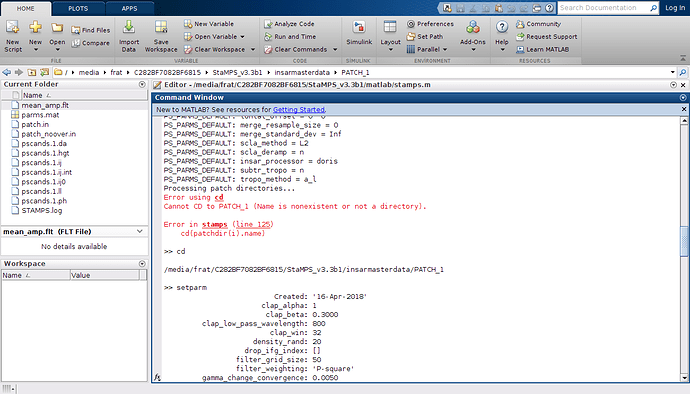
rereading your post and looking on the new screenshots I have an idea what might be your problem. In the Matlab screenshot wit the cd error message, we can see that you are already in the /ubuntustampsexport/PATCH_1 directory and in this directory there is no PATCH_1 folder to enter…
What I assume you have done, after running mt_prep_gamma_snap you have not launch matlab from …/ubuntustampsexport but changed directory to /ubuntustampsexport/PATCH_1 and launched matlab from here. Am I right with this? If so, you have just to launch matlab from …/ubuntustampsexport try:
open terminal in …/ubuntustampsexport
source your Stamps_CONFIGURATION.bash
you do not have to do a new mt_prep_gamma_snap I think
matlab to call Matlab in this (…/ubuntustampsexport) directory
in matlab try to do step 1 with stamps(1,1)
Does this work? Good Luck!
PS: in your Stamps installation folder are some folders and files which do not belong there…I do not think they do any harm, but if you want, tidy up your folder to look like this:
Hello @thho
you mean , ı should work matlab from …/ubuntustampsexport directory ? But ı can only work matlab from the directory which ı installed matlab (from this directory “/media/frat/C282BF7082BF6815/Matlab2017a/bin/glnxa64”)
I mean , even ı add path of the data way of matlab/bin/glnza64 , ı can not work matlab while opening terminal from another directory
I see…but that is exactly what you need to do. Let’s try some things:
- open a terminal in and cd to your home and call matlab from here, in terminal:
cd ~
matlab
does Matlab launch?
- close Matlab and in this terminal cd to your folder with the Stamps Export data you want to process (I recommend to delete your old data and make an new unprocessed INSAR_master_date with StaMPSExportOp from SNAP before trying this). Then call your StaMPS_CONFIG.bash (have you adapt this to your PATH Environment?) :
cd ~/…/INSAR_master_date
source ~/…/StaMPS_v3.3b1/StaMPS_CONFIG.bash
- now you can call mt_prep_gamma_snap again:
mt_prep_gamma_snap
this will process the data as you already had done but in the new export you have to do it again.
- now you should be able to launch Matlab from this folder:
matlab
this will open Matlab.
Please try this and if you do not have adapted StaMPS_CONFIG.bash, please have a look here how to do this.
Hello @thho
Thank you so much but ı still have problem,
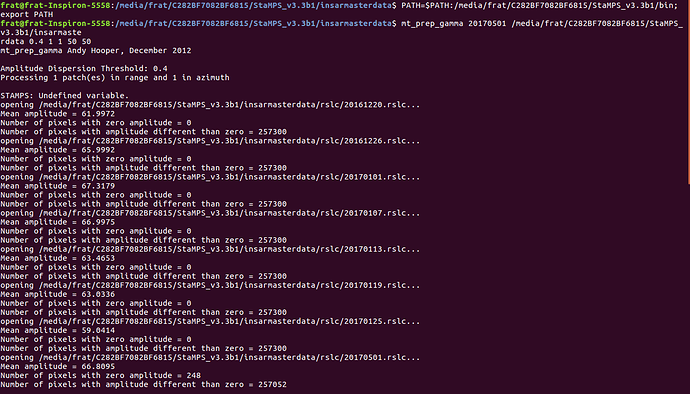
In the picture which ı show you below, like you advised me, ı do the steps; first ı created another Stamps export data in another place inside the Stamps folder, and then ı add path directory way of the stamps/bin into this terminal for working the mt_prep_gamma, and then ı work it and then
like seing at the bottom of 2. terminal picture, ı work matlab from the same terminal without openning another terminal
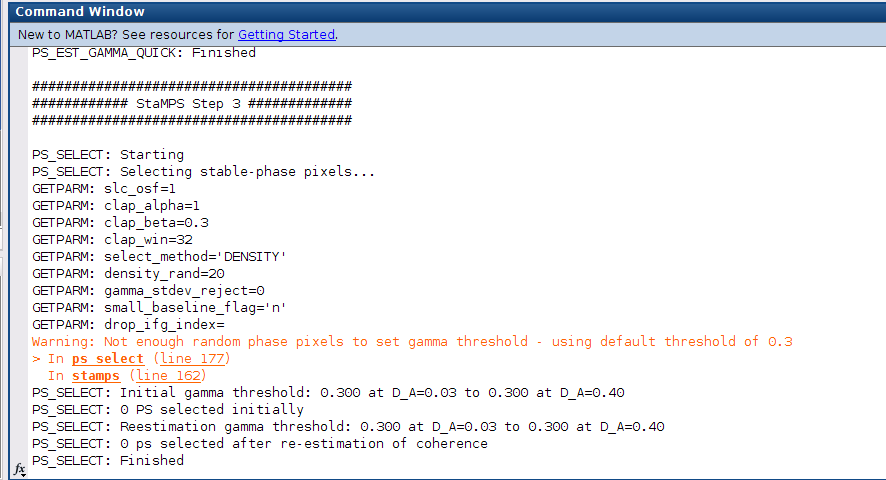
and then on 3. picture ı wrote stamps but it gives me again the same error,
And also, while saying adapting StaMPS_CONFIG.bash , do you mean to add path of this StaMPS_CONFIG.bash directory in to the terminal ? like this (PATH=$PATH:/media/frat/C282BF7082BF6815/StaMPS_v3.3b1/StaMPS_CONFIG.bash; export PATH) ? can you explain me more detailly please ? After adding path of StaMPS_CONFIG.bash, when ı write in the terminal (cd /media/frat/C282BF7082BF6815/StaMPS_v3.3b1/StaMPS_CONFIG.bash), terminal says to me: it is not a directory
the name of your stamPS export directory MUST BE “INSAR_master_data”
Hello @daniel_bona
Thank you so much ,
after fixing the export folder name like you wrote , now ı have another problem like below
what should ı do ?
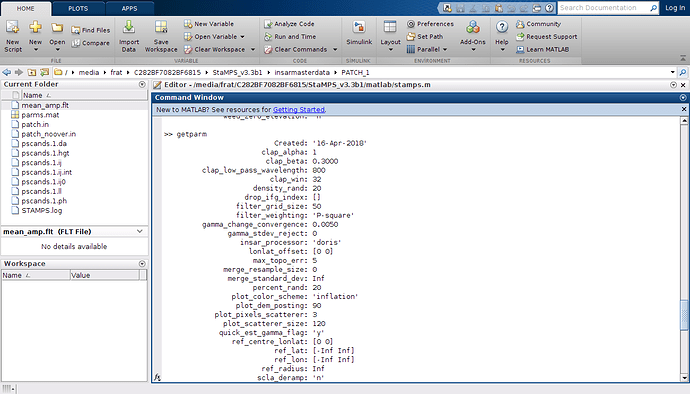
Hi @firat
you are almost there, Just set you Insar Processor from Doris to Gamma by typing this before you run stamps. You should also set your scla_deramp to y by typing second commend before running stamps.
setparm(‘insar_processor’, ‘gamma’)
setparm(‘scla_deramp’, ‘y’)
Have fun
Hello @bayzidul
Thank you so much
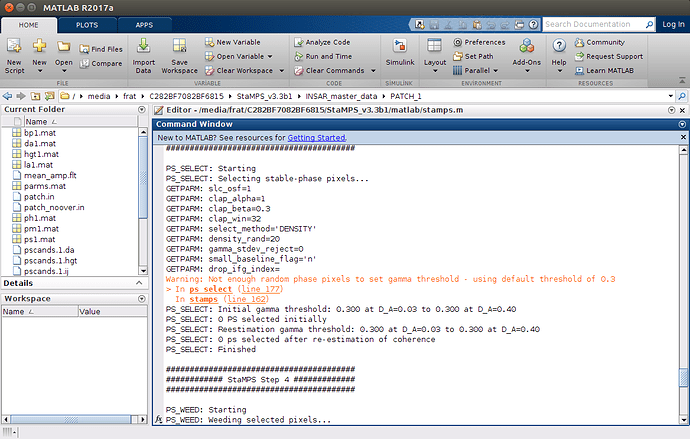
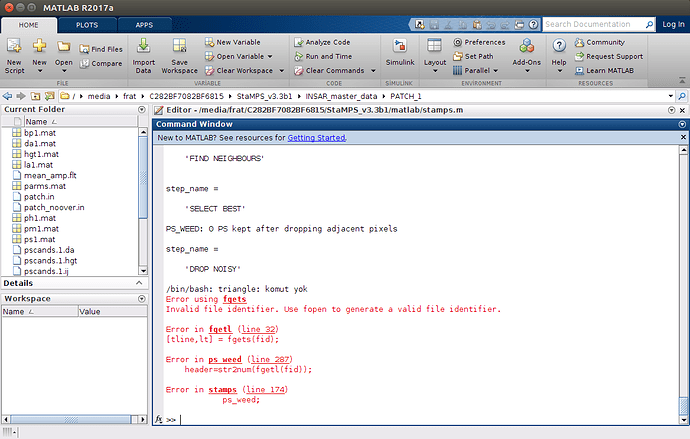
After doing setparm(‘insar_processor’, ‘gamma’) and setparm(‘scla_deramp’, ‘y’), ı have a warning problem on step3 and an error on step 4 which look like below
what should ı do now for processing or passing these steps without any problem ?
Hi @firat,
the name must nor be exactly INSAR_master_date, but INSAR_20160425 if your master image has the date 2016 04 25 (example). I am a bit sceptical if all went correctly, because the processor should be set automatically to gamma, but maybe I have missed that setting the processor with setparm will work to. However, the warning you encountered Not enough random phase may come from two possibilities
-
Your studysite is to small, with setparm(‘filter_grid_size’, 10) I am able to run the script for small areas. But I would recommend to work with larger areas.
-
You can also try to set the dispersion index to a higher threshold, in mt_prep_gamma_snap 0.45 for example, more PS candidates will be preselected
have a try with filter_grid_size or a larger subset of your studysite and let us know if this fixed your problem or not.
Good luck!
Hello @thho
Thank you so much,
After writing setparm(‘filter_grid_size’, 10), there is no warning or error on step 3
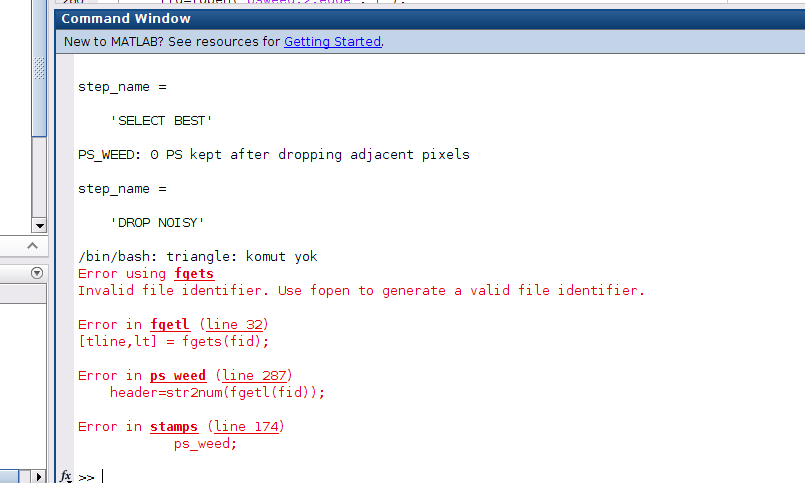
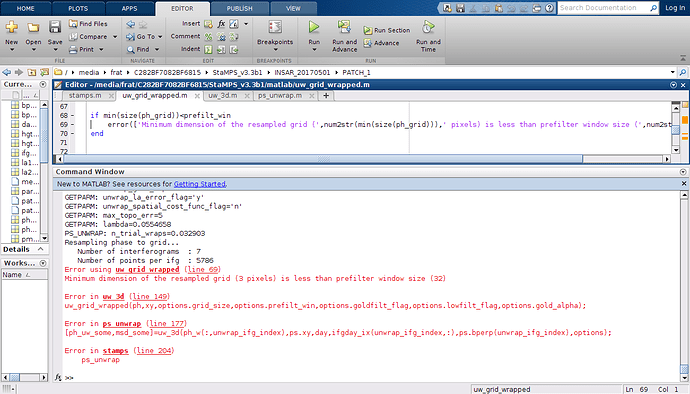
But now ı have errors on step 6 like below
what should ı do for fixing it ?
In step 5 you have to change the parameter ‘merge_resample_size’ at least as high as the ‘unwrap_grid_size’ you want to use in step 6. Think about you want to unwrap with a grid size which is smaller than a resampled grid as input, that is not possible, the unwrapping parameter would be dominated by the merge parameter and therefore useless. Use a unwrap_grid_size of 32 or lager (in your case) or set merge_resample_size to 0 for a first try ( you have to rerun step 5 before this comes to effect)…
I hope this was helpful.
Cheers.