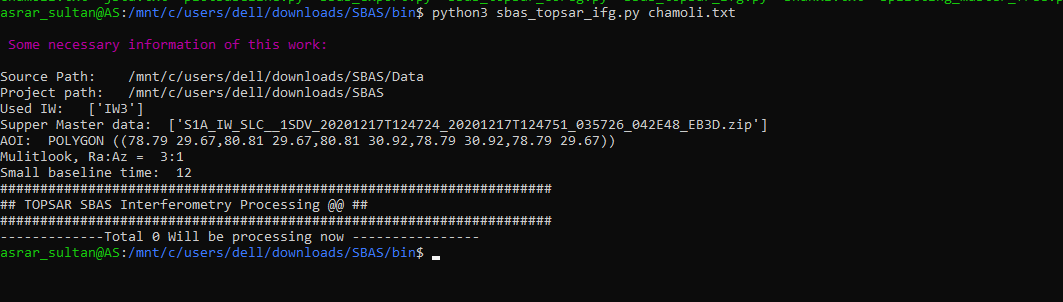
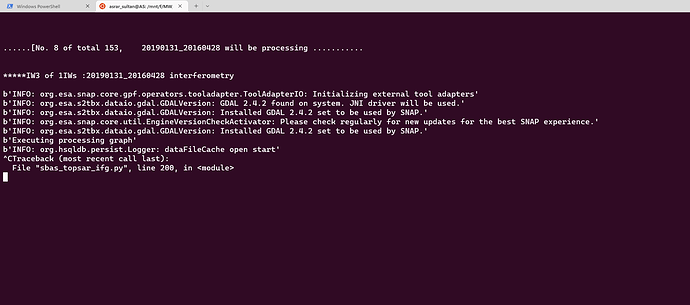
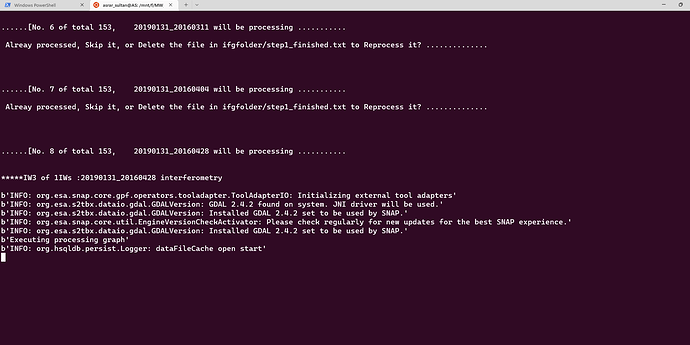
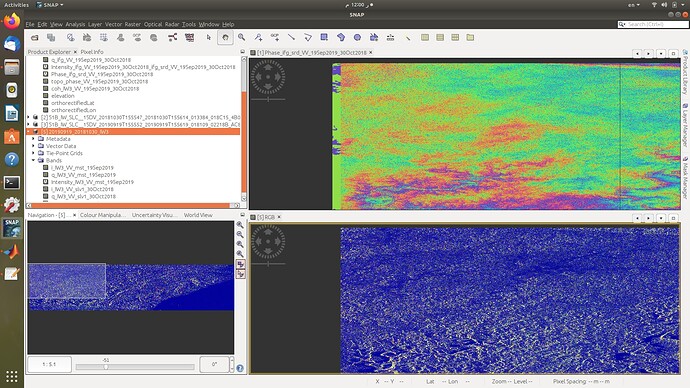
Thank you. Your solution worked perfectly. Now I am getting this error. I dont seem to find such an error in this topic. Could it be due to the fact that python script is unable to find the coregistered files in tempcoreg folder?
Thank you @ABraun. Ifg step is working fine now. Seems to have been an issue of Temp_baseline which I have increased to 144 from previous 12
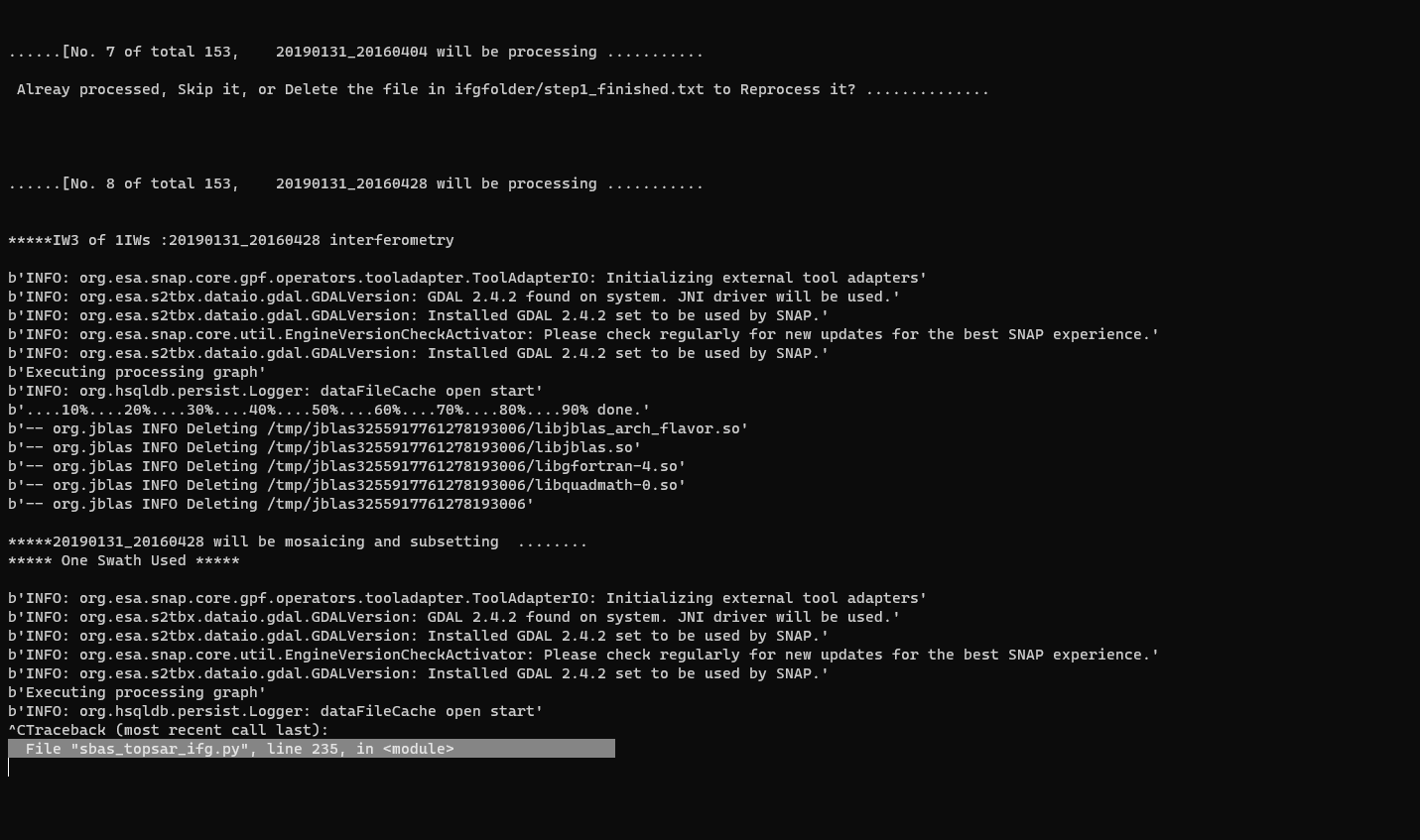
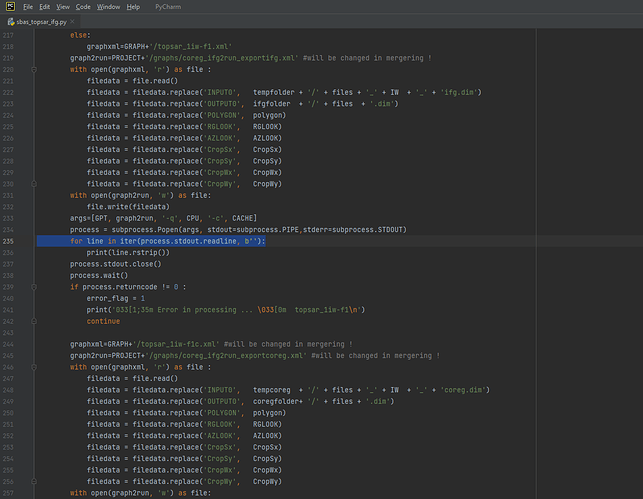
Dear @ABraun, I ran SBAS_Snap2stamps end to end on several datasets for testing and learning purposes and it went smoothly. Then I tried running it for a larger dataset ranging several years and it went smoothly again until sbas_topsar_ifg.py. Here the script worked on a single temp_coreg pair then got stuck at this error:
b’INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 2.4.2 found on system. JNI driver will be used.’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.4.2 set to be used by SNAP.’
b’INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.4.2 set to be used by SNAP.’
b’Executing processing graph’
b’INFO: org.hsqldb.persist.Logger: dataFileCache open start’
I tried changing temporal baseline several times, but didnt solve this issue of getting stuck for ever. I had to rerun the python script several times to even complete a single IFG and COREG pair. The resultant IFGs and COREGs are good in quality. Whenever I try to terminate the process, it always shows Line 200 in module or Line 235 in module. Kindly take a look at this for few moments from your very precious time. Thanks.
Sorry for not solving, I was wondering if you have found a solution for this issue yet?
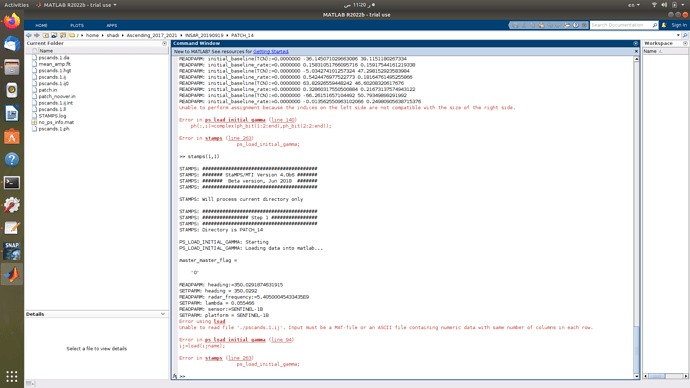
I have seen matlab related issues in mathworks forum and they said that error appears when trying to store a larger matrix into smaller ones regarding their x and y spaces. So when we consider snap-to-stamps situation, It is just maybe related to the export parameters like the number of patches in range/azimuth or the overlapping pixels between patches in either range or azimuth direction. Because the loading and initial selection of PS points worked flawlessly in some patches untill it reached patch 13 out of 21 where this issue appeared
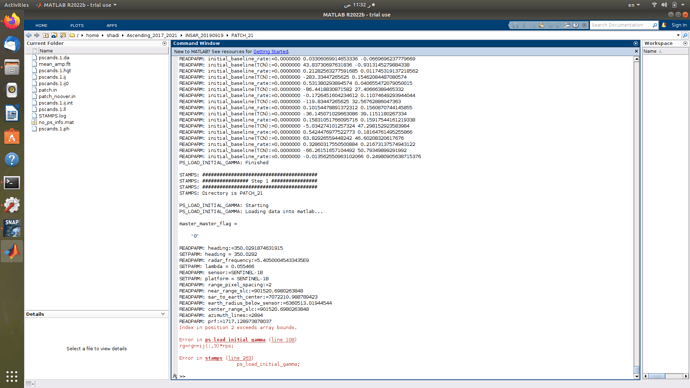
However loading the next patches 15 and 16 till the end seemed to have worked just fine, there’s a problem with the two consecutive patches 13 and 14
and this error message appears during step1 for the next patch 14
[Unable to read file ‘./pscands.1.ij’. Input must be a MAT-file or an ASCII file containing numeric data with same number of columns in each row.
Error in ps_load_initial_gamma (line 94)
ij=load(ijname);
Error in stamps (line 263)
ps_load_initial_gamma;
Just a text copy of the error message to facilitate terms search across the forum
The last patch loading, gave this error,
Index in position 2 exceeds array bounds.
Error in ps_load_initial_gamma (line 108)
rg=rgn+ij(:,3)*rps;
Error in stamps (line 263)
ps_load_initial_gamma;
I may try to re-do the entire export process but I am afraid the data set is large and it would take over 2 days exporting on my slow machine, I wanted to know if there’s an alternative solution ?
I highly doubt it’s export related issues, because I made sure there are no empty interferograms but the sides of the interferograms were like this. There seems to be an additional uncoregistered area that wasn’t properly subset from the master, even though it doesn’t show up in the R-G-B mst-slv-mst pairs. I don’t know if it’s relavent
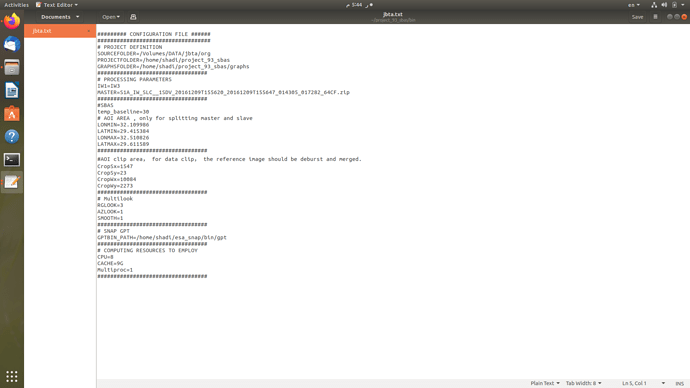
This is my config file, I hope if you can guide to how to edit some lines I am not sure about, such as
SOURCEFOLDER and what’s the difference between it and PROJECTFOLDER? Does the former indicates the path to the input images and the latter is a clean path for the output products? If so, Can I make the both inside the same folder ?
temp_baseline
CropSx=
CropSy=
CropWx=
CropWy=
@Linya-Peng I was hoping if you may take a look at this
Also, Based on what, is the value written for the temp_baseline inside the project config file? Is it the minimum baseline between the consecutive pairs of the input scenes?
Can you share a little bit more explanation to that sentence?
This is what Usai 2001 said but I don’t quite understand clearly what is highlighted in yellow and purple.
Hello Linya,
Thank you so much for your endeavours to solve this intersection issue, I had the same problem you had, and I was really struggling to solve it. And I think you got the right intuition about it when you changed it manually !
To automate it, if you added in the submasters processing in sbas_topsar_coreg.py a simple “if NCOLS in msd_line” and you dealt with it the same way that Band_Raster_Height for instance, the computation would eventually work. Or at least it did for me !
Best regards,
Hello Everybody,
Greetings !!!
I am new to SBAS_InSAR technique. So, I would like to request for help from the Forum regarding SBAS_InSAR processing. Earlier, I have tried to perform PSI using SNAP2StaMPS, StaMPS operation to perform Time-Series Deformation Analysis with the support of RUS-Copernicus tutorial videos. I am using Windows Sub-System for Linux (WSL) in Windows 11 to perform SNAP2StaMPS, StaMPS operation earlier. I am currently using SNAP 9.0.0 version, StaMPS-4.1-beta, snaphu v2.0.4, Matlab R2023b. I am unaware how to start with SBAS_InSAR processing. Can Anyone give me some idea where to start with. If anyone has prepared the SBAS Processing Manual or related videos. I would be grateful to learn from it.
At last i am sorry if it silly things to ask in such Forum.
Diwakar
you can use ASF pipeline
you can use their pipeline on your computer or online with permission…
then there is the yumorishita project with COMET product COMET-LiCS-portal - Centre for the Observation and Modelling of Earthquakes, Volcanoes and Tectonics
GitHub - yumorishita/LiCSBAS: LiCSBAS: InSAR time series analysis package using LiCSAR products
@MarcoSal : Thank you for the suggestion. I will go through yours suggestion and will get back to you if any future quarries arises.
Good morning, first congratulations, good results!
I am wanting to start with the SBAS, would you have a guide or something that can help me please, I would be very grateful.
İf you want to start SABS you can use licsar products with LiCSBAS. İt is very easy to use. Please check GitHub - yumorishita/LiCSBAS: LiCSBAS: InSAR time series analysis package using LiCSAR products. And you can find to videos about licsbas in youtube.