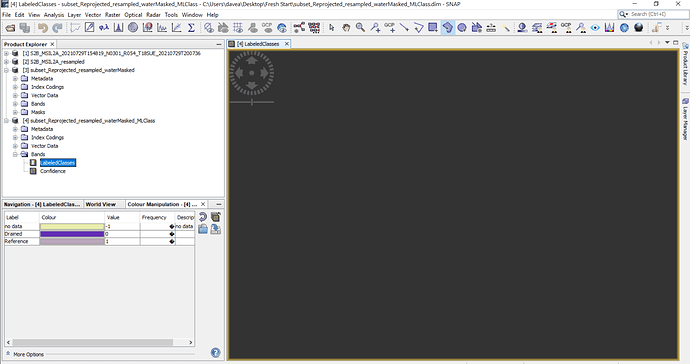
I have reached a point of total desperation: Trying to do a simple supervised Max Likelihood classification
S2B_MSIL2A_20210729T154819_N0301_R054_T18SUE_20210729T200736 - Copy
SNAP 8
Steps
Re-project to WGS84 (Save)
Resample to Band 2 (Save)
Subset (Save)
Create Vector Containers for each class on subset image
-Select all classes
-select all bands
Fail fail fail empty gray space every time but classes show up in color panel. Even after having success once it cannot replicate again. Why on earth wouldn Snap just be designed to use the given projection as the images it creates
Run classification