Could you show your graph?
graph_resample_subset_geotiff_STEP.xml (2.4 KB)
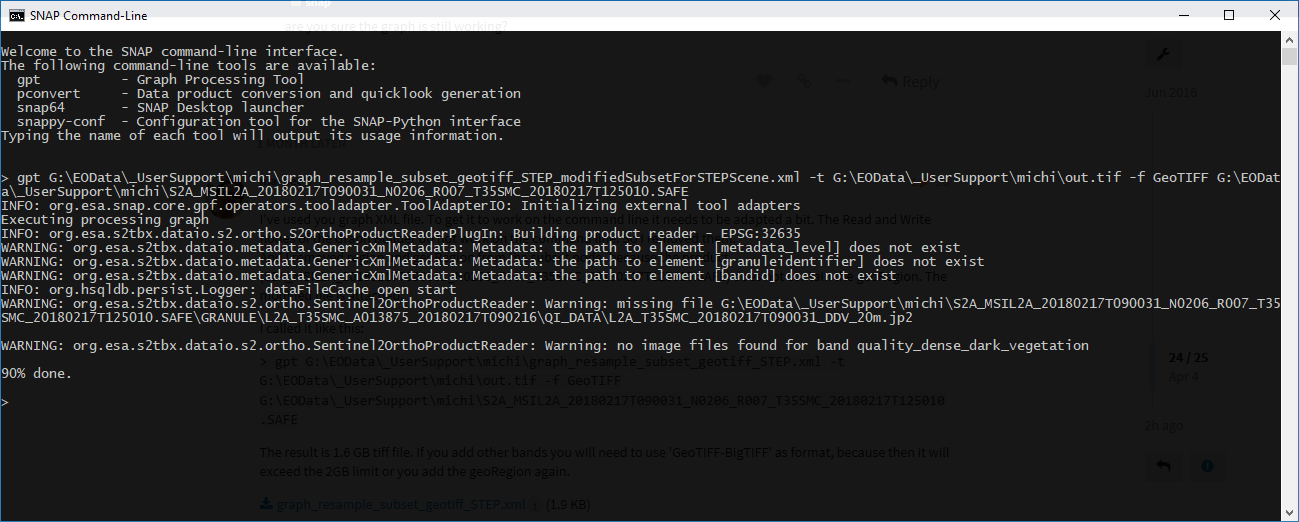
the attached graph is working if I run it in SNAP 6.0 desktop application (but the band subset is not working?). If I run it via gpt I get the Error from above…
Thank you for helping!
Hi,
I have tested your graph and it seems to be working properly here. Could you please execute the command “gpt --diag” to verify that the version is also the 6.0 when you execute gpt? Perhaps you have an old version of SNAP installed and your path is not updated.
gpt --diag
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing ex
ternal tool adapters
SNAP Release version 6.0
SNAP home: C:\Program Files\snap\bin/…
SNAP debug: null
SNAP log level: null
Java home: c:\program files\snap\jre
Java version: 1.8.0_102
Processors: 16
Max memory: 19.6 GB
Cache size: 1024.0 MB
Tile parallelism: 16
Tile size: 512 x 512 pixels
To configure your gpt memory usage:
Edit snap/bin/gpt.vmoptions
To configure your gpt cache size and parallelism:
Edit .snap/etc/snap.properties or gpt -c ${cachesize-in-GB}G -q ${parallelism}
any other ideas?
Which product do you try to resample? Can you post the name? Have you tried to use DIMAP as output format? Is this working and is the result multi-size?
I remember there was some problem when storing resampled S2 to GeoTiff. But this should have been fixed in version 6.
have you tried the graph I provided here:
This once worked. In the Graph file you can switch to Big-GeoTiff format.
In the header of the graph is an example call shown.
Fist of all: thank you very much for helping!
To you questions:
-
Which product do you try to resample? Can you post the name?
S2A_MSIL1C_20170704T112111_N0205_R037_T29TPE_20170704T112431.SAFE -
Have you tried to use DIMAP as output format? Is this working and is the result multi-size?
I remember there was some problem when storing resampled S2 to GeoTiff. But this should have been fixed in version 6.
Yes I tried this and it is working and properly resampling all bands to 10m (from gpt as well as from Destop SNAP). I could do a workaround with the DIMAP-product, but than I have to clean up afterwards and the strange thing that my gpt is working differently than the Desktop SNAP is still not resolved…??? -
have you tried the graph I provided here:
I could not download the product you used (S2A_OPER_PRD_MSIL1C_PDMC_20160414T041702_R008_V20150812T104021_20150812T104021.SAFE is not available anymore), so I used S2A_MSIL1C_20180227T104021_N0206_R008_T31TDH_20180227T141612.SAFE which is in the same area.
- the graph did not work in SNAP desktop -> Error: invalid sub-sampling
- via command line I got the same multisize-error like with my graph
are you sure the graph is still working?
I’ve used you graph XML file. To get it to work on the command line it needs to be adapted a bit. The Read and Write nodes of the Graph Builder do not work on the command line. So I removed them.
And I removed region and geoRegion from the subset node, because the product (S2A_MSIL2A_20180217T090031_N0206_R007_T35SMC_20180217T125010.SAFE) does not contain the geoRegion. The modified file is attached.
I called it like this:
> gpt G:\EOData\_UserSupport\michi\graph_resample_subset_geotiff_STEP.xml -t G:\EOData\_UserSupport\michi\out.tif -f GeoTIFF G:\EOData\_UserSupport\michi\S2A_MSIL2A_20180217T090031_N0206_R007_T35SMC_20180217T125010.SAFE
The result is 1.6 GB tiff file. If you add other bands you will need to use ‘GeoTIFF-BigTIFF’ as format, because then it will exceed the 2GB limit or you add the geoRegion again.
Thank you very much for your help!
your graph and call via command line works for me too. But my problem still remains: if I add a geoRegion (modified to the scene you used) I get the error: “Error: [NodeId: WriteOp@Subset] Product writer is unable to write this product as ‘GeoTIFF’: Cannot write multisize products. Consider resampling the product first.”
I attached the modified xml file. graph_resample_subset_geotiff_STEP_modifiedSubsetForSTEPScene.xml (1.9 KB) Maybe you can check if this works for you…?
Yes, it worked. Also with your graph.
Have you tried to reinstall SNAP already?
When uninstalling, chose to remove all data, not only the SNAP internal data.
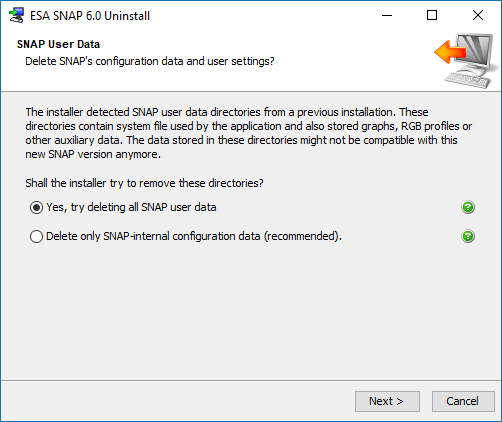
Maybe there is something left over from a previous installation.
i did, still the same error…
maybe there are some updates missing, if i try to update the server is not available:
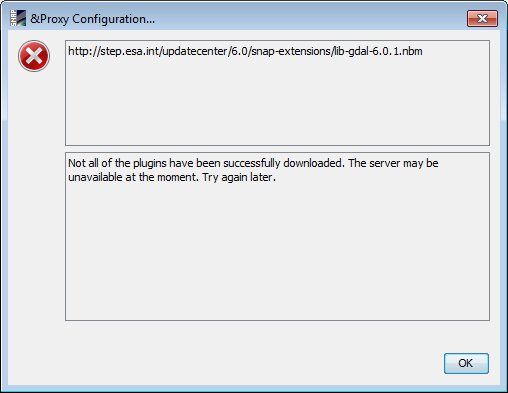
Maybe this happend because of a our server outage. We struggle with such outages in last weeks.
The file exists on the server. If you try it again, it should work. If the server is alive.
finally it works again! after uninstalling/installing several times, i got it working by also deleting C:/Users/user/.snap, C:/Users/user/Favorites/SNAP, C:/Users/user/AppData/Roaming/SNAP before reinstalling.
Thank you so much for helping!!!
Hi i would like to ask. How to do layer stacking in SNAP? Also, how to do resampling in SNAP, i’ve been trying to do it so many times, and it shows an error when i tried to save as Geotiff. I save it in Geotiff because later I have to open it on ENVI.
especially when you want to open it in ENVI, you can stick to the BEAM DIMAP product. It writes ENVI files inside the .data folder of your output location.
Each BEAM DIMAP product consists of .dim and .data
inside the .data folder:
You can use the Collocation tool which allows you to define one master image and a resampling method which resamples all slave products (the other bands in the stack) according to the spatial dimensions and resolution of the master band.
How about layer stacking only on sentinel 20m images only? Do i have to perform resample before doing collocations? 
why would you need resampling if they all have 20 m?
I’m so sorry but I don’t really understand. I’m working on sentinal 2a images. The files contains resolution images of 10m, 20m and 60m. I want to do a layer stacking process on 20m, later on i need to open it in ENVI for pre-processing.
sorry, I thought you had already prepared the images.
If you want to include all S2 rasters, you have to run the S2 resampling operator first, yes. Afterwards, you can use the outputs in the Collocation tool.
If you only want to use the bands with 20 m resolution, you can use the Band Select operator to select those of interest and afterwards use them as input for the Collocation tool.
Thank you!