Yes, this should be enough.
Can you show me the configuration you have used?
Which product do you try to resample, can you give me the name?
Are you working on the command line or in the desktop application?
Last question, which operating OS are you using?
The configuration that I have used is on the image below:
The name of the image that I try to resample is:
S2A_OPER_MTD_SAFL1C_PDMC_20160429T163731_R093_V20150818T092006_20150818T092006
I work in the desktop application, did you suggest to work on the comand line?
I am using windows10 x64 bit
Working on the command line or in the desktop application should not matter. I’ll try to replicate your use case tomorrow.
We just tried it with the same product also on a PC (Windows10 x64) which has 16GB RAM too. It worked well.
Some other reason why it might fail on your machine:
- You have increased the memory after the SNAP installation
- You have other memory consuming tasks running in the background
- You haven’t installed all modules updates
I could not update plugin installer, and it always remind me unconnected to updata center. I want to know how to resample sentinel2A data in a shapefile extent and in band 2,3,4,8. I appreciate for your reply.
Maybe you are behind a proxy server which causes problems or there is some other problem why you can not connect to our server. I once heard that the “Great Wall” caused problems. Maybe you should talk to your IT admins.
Have you followed this thread:
For the resampling, as long as you don’t have the latest modules it might be problematic to resample S2 data.
But in general you can do it.
Using a shapefile to crop the data to a certain area is not easy. In the GUI you can define a geographic bounding box and from the command line you can specify a polygon using WKT. But this will cut out also only the bounding box of the polygon (because images have to be rectangular). You can mask the data afterwards by using the land/sea mask processor.
You will find several examples in the forum how to do this.
Hi,
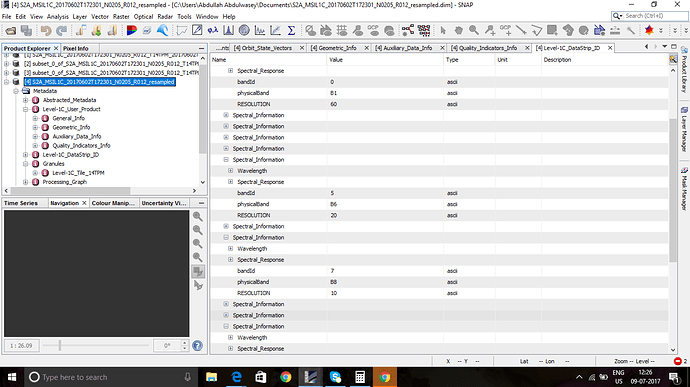
I tried to resample a S2A image, and after that I tried to compute biophysical indices LAI and APAR and got errors. When, after resampling, I checked the meta data, all the bands shows native resolutions. Please see the screen shot. Why resample did not converted all the spatial resolution to 10 m , any idea? Please help. Thanks.
The image that I had downloaded is S2A_MSIL1C_20170526T172901_N0205_R055_T14TLL_20170526T173505
The metadata information is not changes during resampling.
You can check the size of a band by selecting it and the open the Information Window ![]() .
.
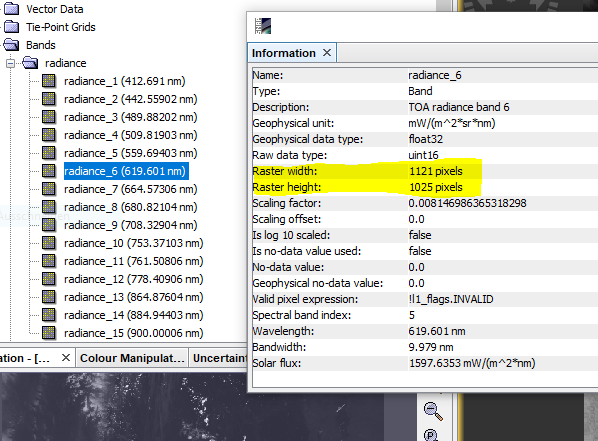
My main issue is I was not able to compute LAI and fAPAR and the error was the S2MSI bands have different spatial resolution. To make all the bands equal spatial resolution of 10 m I tried to re sample it, but it did not work. How should I overcome this problem and compute LAI?
thanks!
I now see the problem: When performing LAI on a Sentinel-2 image an error message is displayed which is different from the one which is directly leading to the resampling dialogue:
LAI /fPAR
other tools (e.g. subset)
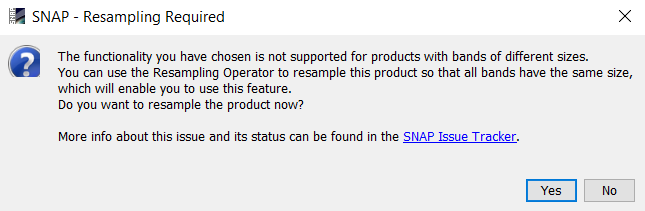
It would be good if any tool led to the second one so resampling could be directly accessed by clicking on “Yes”
@Abdulwasey: You can manually start the resampling tool with Raster > Geometric Operations > Resampling
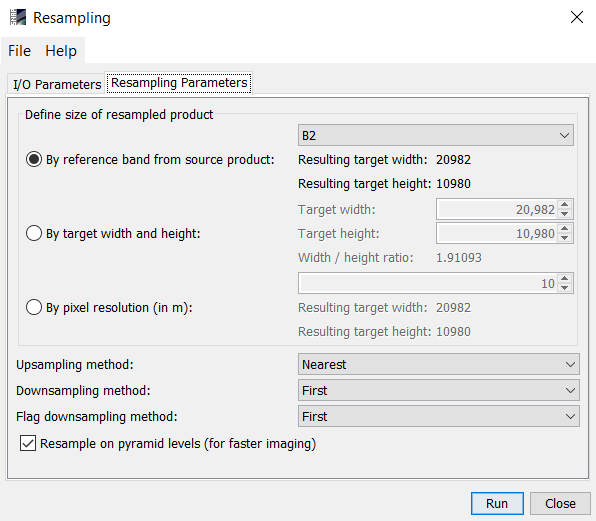
Here you can define a common resolution for all bands (e.g. 10m).
the above image i can see that the resampling size is define by the pixel size of B2 which is 10m
but if i want to define the resampling by pixel resolution then what i have to do
then you should select “by pixel resolution (in m)”
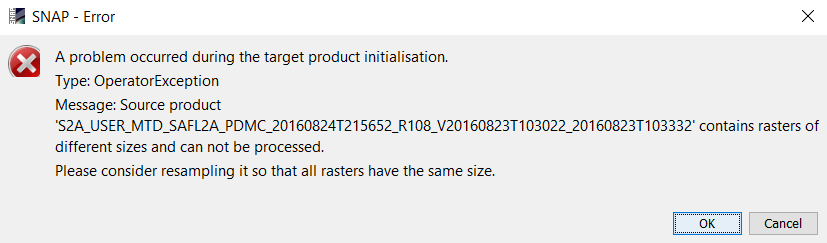
I have exactly the problem, that the my “resampling/subsetting/export to Geotiff”-Graph is working if I run it via the Graph Builder in the SNAP 6.0 desktop application, but if I run the same Graph via commandline using gpt I get an error
Error: [NodeId: Write] Product writer is unable to write this product as ‘GeoTIFF-BigTIFF’: Cannot write multisize products. Consider resampling the product first.
Any tips?? Thanks
Could you show your graph?
graph_resample_subset_geotiff_STEP.xml (2.4 KB)
the attached graph is working if I run it in SNAP 6.0 desktop application (but the band subset is not working?). If I run it via gpt I get the Error from above…
Thank you for helping!
Hi,
I have tested your graph and it seems to be working properly here. Could you please execute the command “gpt --diag” to verify that the version is also the 6.0 when you execute gpt? Perhaps you have an old version of SNAP installed and your path is not updated.
gpt --diag
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing ex
ternal tool adapters
SNAP Release version 6.0
SNAP home: C:\Program Files\snap\bin/…
SNAP debug: null
SNAP log level: null
Java home: c:\program files\snap\jre
Java version: 1.8.0_102
Processors: 16
Max memory: 19.6 GB
Cache size: 1024.0 MB
Tile parallelism: 16
Tile size: 512 x 512 pixels
To configure your gpt memory usage:
Edit snap/bin/gpt.vmoptions
To configure your gpt cache size and parallelism:
Edit .snap/etc/snap.properties or gpt -c ${cachesize-in-GB}G -q ${parallelism}
any other ideas?
Which product do you try to resample? Can you post the name? Have you tried to use DIMAP as output format? Is this working and is the result multi-size?
I remember there was some problem when storing resampled S2 to GeoTiff. But this should have been fixed in version 6.
have you tried the graph I provided here:
This once worked. In the Graph file you can switch to Big-GeoTiff format.
In the header of the graph is an example call shown.
Fist of all: thank you very much for helping!
To you questions:
-
Which product do you try to resample? Can you post the name?
S2A_MSIL1C_20170704T112111_N0205_R037_T29TPE_20170704T112431.SAFE -
Have you tried to use DIMAP as output format? Is this working and is the result multi-size?
I remember there was some problem when storing resampled S2 to GeoTiff. But this should have been fixed in version 6.
Yes I tried this and it is working and properly resampling all bands to 10m (from gpt as well as from Destop SNAP). I could do a workaround with the DIMAP-product, but than I have to clean up afterwards and the strange thing that my gpt is working differently than the Desktop SNAP is still not resolved…??? -
have you tried the graph I provided here:
I could not download the product you used (S2A_OPER_PRD_MSIL1C_PDMC_20160414T041702_R008_V20150812T104021_20150812T104021.SAFE is not available anymore), so I used S2A_MSIL1C_20180227T104021_N0206_R008_T31TDH_20180227T141612.SAFE which is in the same area.
- the graph did not work in SNAP desktop -> Error: invalid sub-sampling
- via command line I got the same multisize-error like with my graph
are you sure the graph is still working?