Thank you!
I used the right file. I will try to process the images again.
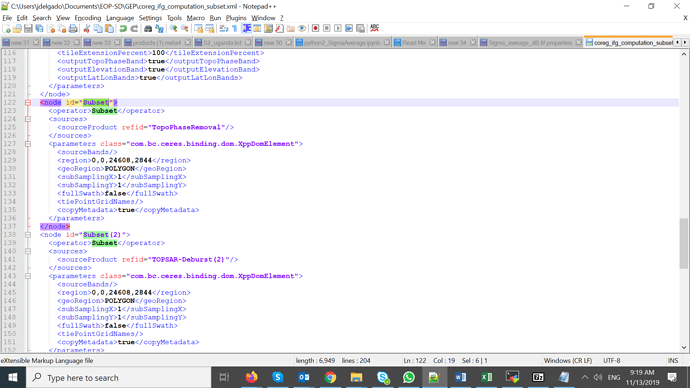
Please @Sharon, be careful while opening the graphs within SNAP 7.0. They were created with SNAP 6.0, so better open them with an editor (vim, gedit, notepad), so you can search the word Subset and you will find Subset and Subset(2) as operators.
I confirm that the coreg_ifg_computation_subset.xml contains the Subset operators needed in the github and zenodo snap2stamps packages.
In case that your graph using SNAP 7 is different, try to reconstruct it yourself.
We can try to support you on that. Just please be careful with statements such as
as may be confusing for other (new) users, when deciding to start using the package.
See in screenshot below the 2 Subsets Operators found in the aforementioned file in lines 122-153
Let us know if you manage to solve your issue or you may need more support
Best regards,
Jose Manuel
If you have issues with memory and heap space, use shorter graphs. For exampkle 1) coregister 2) InSAR operations 3) terrain correect.
this is true, but the python scripts of snap2stamps already call these as separate graphs:
- splitting all slaves
- coregister all slaves with the master
- generate interferograms
- exporting the data to StaMPS format
Terrain correction is not required.
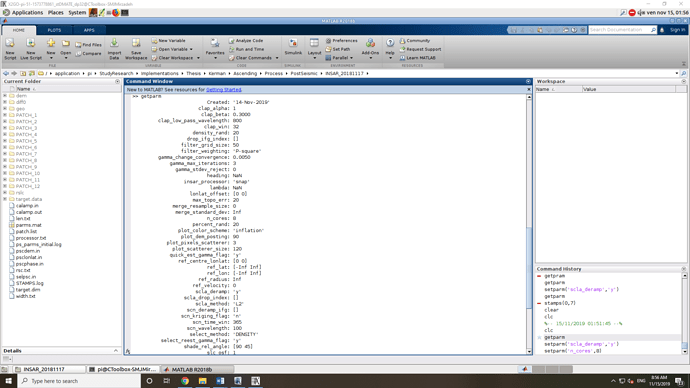
@mdelgado Sorry to disturb you. I just finished the processing with the package and started running the StaMPS.When I checked the parameters using the “getparm” command, I found that the values of “heading” and “lambda” set to “NaN”. I was wondering if it is not true and I will face an error in the following steps. Please let me know it is correct or not.
Best regards,
Sayyed
Is this before applying step 1?
If so… it may be normal. Please check after step1
@mdelgado Thank you for your comment. I checked them and everything is OK.
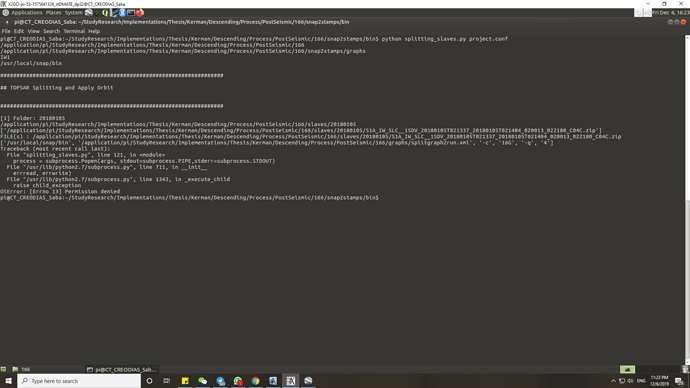
@mdelgado I would like to run the package but I faced the following error:
Could you please guide me on how I can fix it?
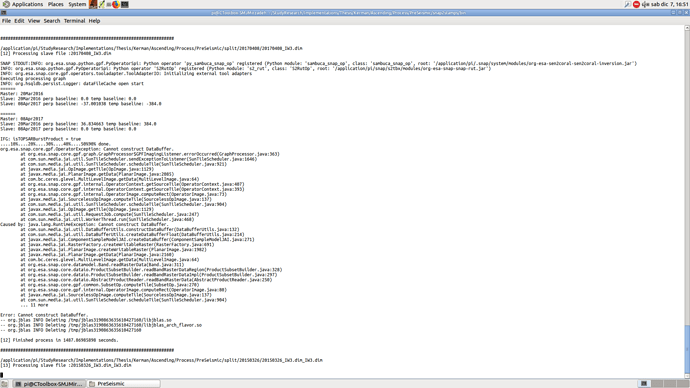
@mdelgado Hi. Excuse me, I checked my progress and found an issue for one of the slaves [12] as follows:
I do really appreciate if you let me know whether I have to re-run this step after finishing the progress?
“Cannot construct Data Buffer“ means the operation was terminated. Probably no correct product was written in this step.
From the first error I would suggest you to check your read/write permissions on the involved folders
I believe that the Cannot contruct data buffer is related to memory. How big is your system memory and how many burst contains your AOI? Had you assembled several slices?
As you can see, the SNAP software was installed in /usr/local. I have permission to this folder to install he software and package but I don’t know why I got this error. Should I run the command by “sudo”?
My system in RSS has 8 CPU and 32 RAM. Yes, I assembled two slices because of the coverage of the images but the number of selected bursts are less than 9 ones.
If I re-run this step, will it create the correct ones again? Or It just will handle the uncompleted slaves?
Well… that error could also for the folders you want to read and write
As it is the first time somebody reporting such error… I believe that it is not related to snap2stamps package itself but on you want them to read and write
The script does not check the integrity of the files found, so it will try to recreate all. You can avoid this by removing from the slave splitting folder the ones already processed.
I hope this helps
Thank you so much. Fortunately, you point worked very well ![]()
Yes, I know. Exactly, it is not related to the package. I will ask the RSS support team of to help me fix it.
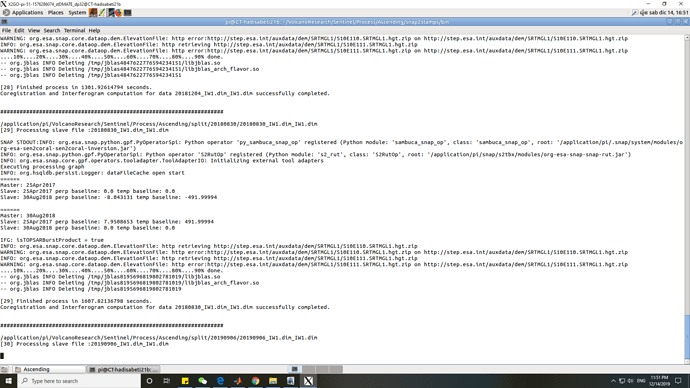
@mdelgado Sorry, I put the package in running and found in the command that the processing is doing successful but something warning about the dem file made me worried. Could you please see the following picture and let me know it is important or not?
you have to check the coregistered products: Open them in SNAP and check if the coregistration was successful as described in this post. Also, check if the included elevation band contains data. If not (it will appear purple only), the removal of topographic phase was not carried out correctly.