Hi there.
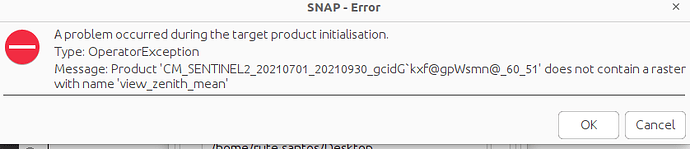
I created a mosaic in python where I used several Sentinel-2 images from both sensors (S2A and S2B) before merging all together in a median mosaic. I know want to use this mosaic in SNAP to run the Biophysical Processor. But when I run I get an error
I thought this could be due to the lack of the metadata file but I cannot uploaded it. Still, considering that we need to select the sensor we are using I am assuming it is not possible to use this mosaic for this method. Am I right?
Thank you
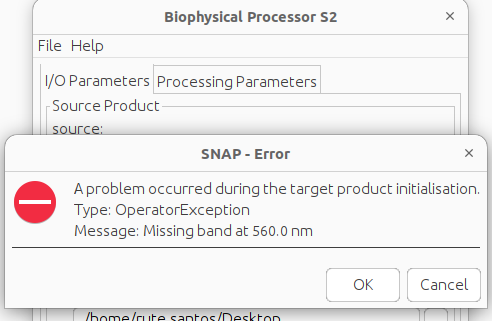
The processor cannot detect the wavelength it needs. You can try to specify them for the bands of your mosaic. Also the band names need to match the expectation.
It might be easier to do first the processing with the biophysical processor and then mosaic the results.
1 Like
Hi, thanks! I did change the names and added the wavelengths, however I get a new error. I understand I do not have this file since I’m working with a mosaic I created. I’m concluding that there’s no way I can run this on the mosaic.
As for the suggestion on running this before the mosaic, this would lead me to run this process on a lot of images which would increase my work. I’ll search for another way to do this outside of SNAP.
Can’t you process with the Biophysical Processor first and make the mosaic at a later step?
No. This mosaic for example is the result of 50 images. That would take me a lot of time if I had to do this to all 50. I’ll see another way.
If you use scripting it should not be that time-consuming or tedious to process 50 scenes.
at the end of the day that is not the question I asked. I simply wanted to know if it was possible to use the biophysical processor on a mosaic.