I’m trying to replicate output of the Radar > Coregistration > Stack Tools > Create Stack. I have some pre-processed Sentinel-1 file stored in the data folder, from which I select file for reference by date. When I create stack using SNAP Desktop in the result I get slave bands in a chronological order and file which is at the top of the list is used as a reference (master).
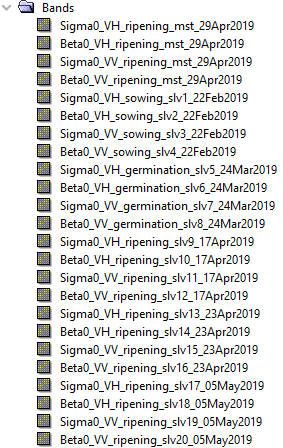
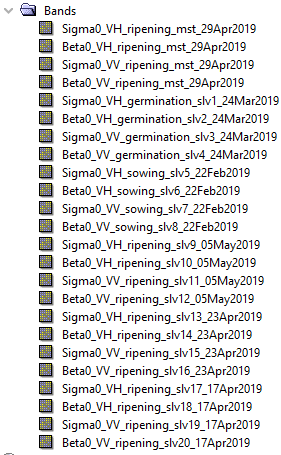
I learned by trial and error that in my script I need to put the reference file at the end of list (products list in the code below) passed to the CreateStack method, but in the output file slave bands are in random order. I adjusted my script to match the process flow which is in the Processing_Graph of the file generated by SNAP Desktop, but I get different output.
I think that similar issue was mentioned in the past in this topic: Call ProductSet-Reader and Create Stack in Snap
I’ll appreciate any suggestions about what I can try to solve this issue, because I’m out of ideas.
Here is my Python code:
import os
HOME_PATH = os.path.expanduser("~")
SNAP_PYTHON_PATH = os.path.join(HOME_PATH, ".snap", "snap-python")
import sys
sys.path.append(SNAP_PYTHON_PATH)
import esa_snappy
import glob
DATA_PATH = "data"
REFERENCE_DATE = "20190429"
OUTPUT_FILE_NAME = "stack_file_script.dim"
files = glob.glob(DATA_PATH + "\*.dim")
files.sort()
# Put reference file as the last one in list
files_reordered = [file for file in files if REFERENCE_DATE not in file] + [ref for ref in files if REFERENCE_DATE in ref]
# Append working directory to file path to match SNAP Desktop behaviour
files_reordered = [os.path.join(os.getcwd(), file) for file in files_reordered]
products = []
print("Files order:")
for file in files_reordered:
parameters = esa_snappy.HashMap()
parameters.put("useAdvancedOptions", "false")
parameters.put("file", file)
parameters.put("formatName", "BEAM-DIMAP")
parameters.put("copyMetadata", "true")
file_data = esa_snappy.GPF.createProduct("Read", parameters, products)
products.append(file_data)
print(f"{products[-1].getName()}")
parameters = esa_snappy.HashMap()
parameters.put("resamplingType", "NEAREST_NEIGHBOUR")
parameters.put("initialOffsetMethod", "Orbit")
parameters.put("extent", "Master")
stack_data = esa_snappy.GPF.createProduct("CreateStack", parameters, products)
file_name = os.path.join(os.getcwd(), OUTPUT_FILE_NAME)
parameters = esa_snappy.HashMap()
parameters.put("writeEntireTileRows", "false")
parameters.put("file", file_name)
parameters.put("deleteOutputOnFailure", "true")
parameters.put("formatName", "BEAM-DIMAP")
parameters.put("clearCacheAfterRowWrite", "false")
stack_data = esa_snappy.GPF.createProduct("Write", parameters, stack_data)
EDIT
It seems I was just lucky with setting the reference file. I added more files to my test folder and it stopped working. It’s not random, because every time I run the script the same file is selected, but it’s the one from inside of the list (it’s neither first, last or middle file on the list which I pass). It may be related to the order of the read processes executed on the Java side to create the products list. In the output file metadata in the Processing_Graph > node.4 I see path to this selected file passed as a value of the file parameter to the Read operator. It seems that SNAP Desktop reads files in order, but script randomises this for whatever reason.