Hi,
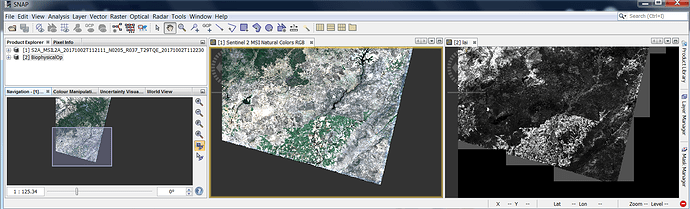
When the biophysical processor is used on an image which has diagonal edges, it creates artifacts around those edges. This is illustrated in the screenshot below:
I used Sen2Cor 2.4.0 for atmospheric correction and SNAP 6.0-PREVIEW5 to derive LAI using a simple graph (Read > Resample > BiophysicalOp > Write). I’m not sure if the issue is new to SNAP 6 or if it has existed in previous versions.
Thanks
Hi,
I have just reproduced these artifacts with version 5. It seems that in those pixels there are values that are not NaN in the angle bands and the processor (a neural network) computes an output with them, but this outputs have no physical sense.
Hi Obarrilero,
Thanks for investigating! I guess this should be quite easy to fix by removing output values for pixels in which all reflectance values are 0 even though angle information is present.
However, there is also an opposite effect (visible on the bottom diagonal edge in the screenshot), where LAI is not estimated even though reflectance values are present. Does this mean that no angular information is available for those pixels?
Hi,
yes, I think that the missing angle values should be the reason for that. I have just created an issue in our issue tracker: https://senbox.atlassian.net/browse/SIITBX-287
Thanks for reporting!