I am unable to process “Biophysical Processor LANDSAT8” which is available in new update of SNAP.
unable to read the input file of Landsat-8.
Plz guide me. Plz tell me which version of Landsat-8 can be readable.
Thank you.
if you download data from the USGS Earth Explorer, you can select Collection 1 Level 1 which is supported by SNAP. I haven’t tested higher level products.
Ok thank you @ABraun I am downloading Landsat-8 collection-1 level-1 data. Till now I tried with collection-2 and level 1,2 dataset. Let’s see.
Thank you for your quick response.
Still biophysical processor for Landsat-8 is not working…
can you please specify “not working”?
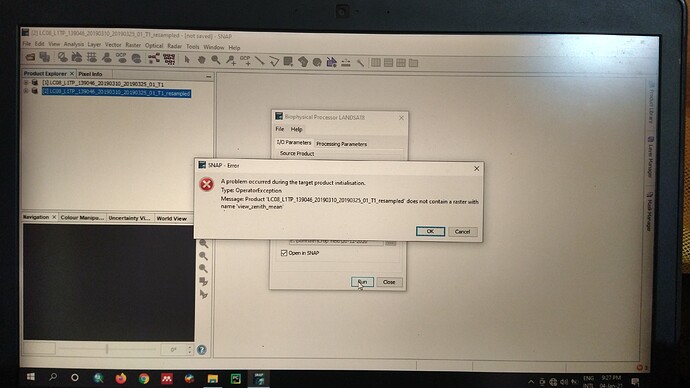
In the biophysical processor I have given the collection 1 and level one dataset of Landsat-8. Then it showed that it should be resampled. I did it. Then when I gave the input in Biophysical Processor for LAI estimation it is showing error that “the input file does not contain a raster with name ‘view Zenith mean’”…
yes, as written in the documentation of this processor:
For the LANDSAT8 the same approach is used, but in this case the bands used as inputs are the blue, red, near_infrared, swir_1 and swir_2 in addition to the auxiliary bands (zenith and azimuth both for sun and view)
That means you have to create the bands
- view_zenith_mean
- view_azimuth_mean
- sun_zenith_mean
- sun_azimuth_mean
with the Band Maths, based on the information you find in the metadata of the product.
.
in the metadata file, it is written that:
SUN_AZIMUTH = 130.2619
SUN_ELEVATION = 54.24
From this information, how will I make the bands which you mentioned above?
open the band maths and create new bands with these names and enter the value in the expression dialogue.
Thank you for your all positive supports…
how will I get “slope-effect corrected reflectances”… As it is mentioned in the manual of Landsat-8 biophysical processor of SNAP, to get optimal results we have to use this slope-effect corrected reflectance…
plz, help.
I am using a product of Landsat-8: LC08_L1TP_139046_20190310_20190325_01_T1
From the metadata file, I have got the geometry information like SUN_AZIMUTH = 130.2619
SUN_ELEVATION = 54.24… But to run the biophysical processor it needs extra three bands and they are:
- view_zenith_mean
- sun_zenith
- view_azimuth_mean
can u plz say these values from the above mentioned two values as I have some confusion? If u say, I can recheck my calculations.
thank you.
I have attached the metadata files…
LC08_L1TP_139046_20190310_20190325_01_T1_MTL.txt (8.4 KB) LC08_L1TP_139046_20190310_20190325_01_T1_ANG.txt (114.5 KB)
@ABraun When I am making the bands like zenith and azimuth in-band math, so all the pixels have the same value. But it should be different…So, for that what should we do ?
these values barely differ within the scene so, if you ask me, using constants is not a bad choice.
Especially because the bands are already called _mean, which assumes an average per image.
Ok, but what would be the value of “view_azimuth_mean” ?
where, SUN_AZIMUTH = 130.2619
SUN_ELEVATION = 54.24 is given…
please have a look here: Generate view angles from metadata Sentinel-2
Hello. Did you solve this problem? Could you please tell me how to get the view_zenith_mean, sun_zenith and view_azimuth_mean from Landsat-8?
Much appreciated.
For Landsat 4-7 and for Landsat 8 there are tools to compute the geometries:
Solar Illumination and Sensor Viewing Angle Coefficient Files (usgs.gov)
The tools are linked on the page.
Maybe @FlorianD can clarify how the Landsat data shall be pre processed to get the required angle bands.
There is also a new ATBD, maybe this helps:
Thank you for your reply, I will give it a try!
Hi all, I thought I would add my question here as I thought that it was similar enough to OP’s question (I apologize if I shouldn’t have resurrected a 1.5 yr old thread - I am actually also having trouble figuring out how to make a post…). I am looking to do the same operation as OP(biophysical processing), but using Landsat 9 imagery in lieu of Landsat 8 imagery. I suspect it is a stretch of an expectation, but would the Landsat 8 biophysical processing tool work on a Landsat 9 (level 1 or 2) product? I cannot seem to find any information online (not just in this forum) on deriving Leaf Area Index (or any other biophysical parameter) using Landsat 9 imagery, despite its similarity to Landsat 8 and that it has now been collecting imagery for almost a year.
Also, I am completely new to using SNAP, but I have been learning quickly due to the excellent documentation available and questions fielded on this forum.
Thank you!