Dear Kerstin,
Thank you very much for your advice.
The Study area is eutrophic and small lake (0.32km2).
Abraun said that the reprojection is needed.
I found an idea, but I don’t know if it is good idea
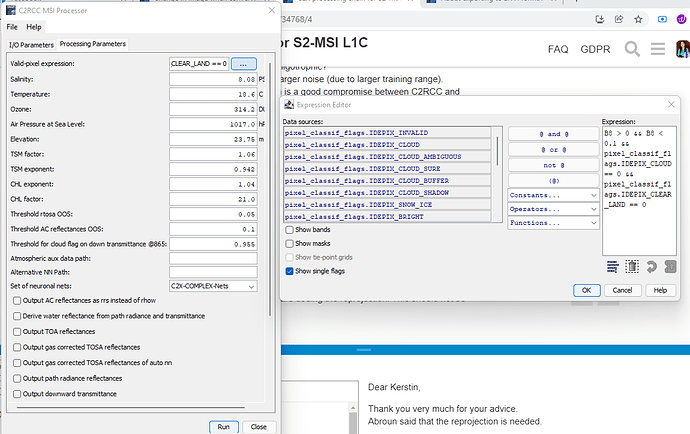
In the Atmospheric correction step, I have applied “B8 > 0 && B8 < 0.1 && pixel_classif_flags.IDEPIX_CLOUD == 0 && pixel_classif_flags.IDEPIX_CLEAR_LAND == 0” this expression as Valid_PE expression. As a result, only remained the clear IDEPIX_CLEAR_WATER.
What do you think about this expression?