Hi everyone!
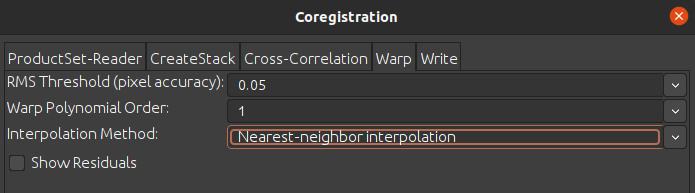
I’m trying to perform Coregistration on a pair of Cosmo-SkyMed second generation (CSG) images. When doing so on SNAP’s GUI I can easily find the right set of parameters for all operators involved (create stack, cross-correlation and warp) to produce a nice pair of correctly overlapped images.
I have to reproduce this operation through the SNAP python-wrapper snapista. I did this until now with no issues for other operations by exporting graphs from SNAP and then setting parameters in python code like so:
g.add_node(
operator=Operator(“Warp”,
rmsThreshold=‘0.05’,
warpPolynomialOrder=‘1’,
interpolationMethod=‘Nearest-neighbor interpolation’,
excludeMaster=“false”,
openResidualsFile=“false”),
node_id=“Warp”,
source=“Cross-Correlation”,
)
In this way, I was able to reproduce the same results from GUI with snapista for every operation except the coregistration: from GUI I get a correct coregistration while with snapista, using the same exact parameters from the exported graph, I get the images shifted by tens of pixels apart.
Now my doubt is, am I missing something by passing parameters to snapista from exported graphs?
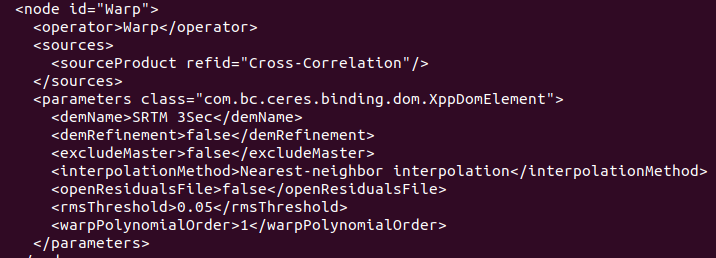
Also, I may have found a hint for this issue, regarding the warp operator: while my code is the one I wrote above, when I print the graph this happens
It appears that automatically sets a DEM, not right for me in this case, while from GUI you cannot even choose one:
The same thing happens with cosmo first generations (CSK) images but the coregistration works fine with snapista too. I don’t know exactly what’s going on here.
Thanks to anyone who took time to read my issue, hope someone helps!