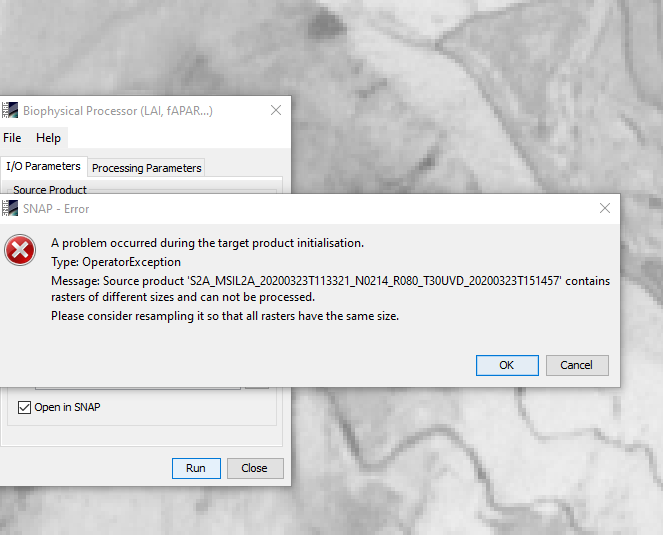
Hi, can anyone help with this error? I’m using the source as downloaded from ESA, so dont understand why it says the rasters are different sizes?
I think you also mentioned a relatively small region of interest, so you may want to work with a spatial subset. Water change detection practical starts with an explanation and example showing how to make bands the same “size”.
Thanks George, I’ll have a look at it
@mattcooper have you resampled all the bands to the same spatial resolution? Not all of the bands that Sentinel-2 captures have the same resolution, so the downloaded images are of different sizes. You can either use a simple resampler, or download the S2 Superres plugin to do this
@BarrowWight, i managed to sort it out in the end, using the method provided by @gnwiii using the graph builder to produce resampled files. One thing that i don’t understand though, when i have done the resampling, how do i open that file in SNAP at a later time, I’ve tried using the ‘open product’ but it doesn’t work!
if you stick to the BEAM DIMAP format, SNAP handles all metadata and you can use “open product”.
Please note that this format consists of one dim-file and one data-folder of the same name. Togehter, they form a product.
Can you be more precise about what “doesn’t work” means. SNAP often delays the actual writing to disk, so if you try to open a newly created file too soon you can get an error or a file that is missing data. The BEAM DIMAP format is generally used for saving intermediate files.
Ah, what i was doing that ‘doesn’t work’ was referring to was clicking ‘open product’ and then trying to select the .data folder in my file explorer window. When i select the .dim file, everything opens fine, it’s just that that file wasn’t visible in the folder view that i had the explorer open in.
very good.
OK, so now I’ve managed to sample a small area of the image, and run the biophysical processor to find the LAI values of each pixel as seen below. Now the questions get more technical, so hopefully this is still in the right place!
All the images aree of the same location, and i am interested to know why there are white areas in the LAI flags image (3rd from left). the patch where the cursor is is grassland, and the white area on the right hand side is very steeply sloping mature coniferous growth. why are these areas flagged? there is a correlation betwen the flagged area and the darkest (lowest) LAI values, but what is the range of the LAI metric, as I cant find it in the documentation.
And the million dollar question, i would like to be able to use these LAI values in a model I’ll be writing in python, is there a way to import the LAI value (indexed by (X,Y) value) into a python script? I’ve managed to do it with .tiff files (from the MODIS product), but snap appears to save the LAi values in a .hdr file which i’m having trouble opening in python.
Being an ocean colour user, I’m not up on the fine details of LAI, but I do use Python.
Ocean colour products have a “flags” band which codes various conditions for which the ocean product generation fails, e.g., land, failures in atmospheric correction, cloud, etc.
There are lots of options for Python. One is snappy, which is a Python library that gives access to many SNAP functions. The GDAL library supports many image formats and can be used from Python. There is also Unidata’s NetCDF4 Python library. NetCDF4-CF has very complete metadata support and has become widely supported by 3rd party tools.
In the spirit of reproducible research, the simplest and most robust configurations are more likely to work the same way for everyone. Snappy is problematic because SNAP includes support for a limited range of Python versions. GDAL is problematic because it has optional support for a large number of file formats but many binary packages omit some formats, and because many packages install private GDAL libraries, each with a different set of supported formats (and bugs). For that reason, Unidata NetCDF is my first choice for exporting SNAP data to be used with 3rd party tools.
Interesting reply @gnwiii, are you suggesting that i export the .hdr file from SNAP as a NetCDF4 file?.
Or is .hdr a format exclusive to SNAP? which seems very parochial to me!
You should be able to save a multi-band product as NetCDF4-CF, but BEAM-DIMAP is useful if you have geometries you want to preserve.
The .hdr files (which are metadata for the associated .img files) are supported by many GIS programs and the GDAL library, but the metadata standardization is not as consistent or complete as with NetCDF4-CF. BEAM DIMAP allows you to load multiple bands in SNAP by selecting one .dim file, but other tools require selecting each .img file. If you also have geometries (feature data --points, paths, etc.) BEAM DIMAP has proprietary support that isn’t compatible with GIS software. NetCDF4-CF can also load multiple bands in SNAP, but GIS software may need you to specify individual bands. GDAL has a special notation “::” that is required when using GDAL libraries (e.g., QGIS).