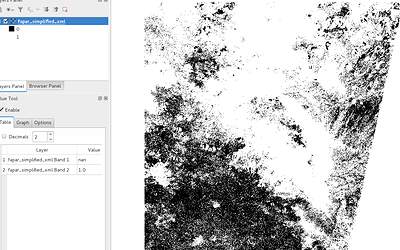
Hello everyone,
Can you help us fixing an issue? We are having very weird results in the computation of fapar (BiophysicalOp), it only outputs NAN results with the following quality flag:
This is the graph we use, input of this graph is the result of Sen2Cor:
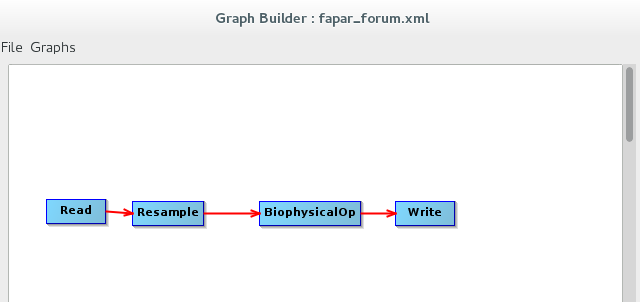
Or in xml format:
<graph id="Graph">
<version>1.0</version>
<node id="Read">
<operator>Read</operator>
<sources/>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<file>/home/dial/nico/copiedProducts/test/S2C_FRW/S2A_MSIL2A_20170214T080021_N0204_R035_T36NXK_20170214T081513.SAFE/MTD_MSIL2A.xml</file>
</parameters>
</node>
<node id="Resample">
<operator>Resample</operator>
<sources>
<sourceProduct refid="Read"/>
</sources>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<referenceBand/>
<targetWidth/>
<targetHeight/>
<targetResolution>60</targetResolution>
<upsampling>Nearest</upsampling>
<downsampling>First</downsampling>
<flagDownsampling>First</flagDownsampling>
<resampleOnPyramidLevels>true</resampleOnPyramidLevels>
</parameters>
</node>
<node id="BiophysicalOp">
<operator>BiophysicalOp</operator>
<sources>
<sourceProduct refid="Resample"/>
</sources>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<computeLAI>false</computeLAI>
<computeFapar>true</computeFapar>
<computeFcover>false</computeFcover>
<computeCab>false</computeCab>
<computeCw>false</computeCw>
</parameters>
</node>
<node id="Write">
<operator>Write</operator>
<sources>
<sourceProduct refid="BiophysicalOp"/>
</sources>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<file>/home/dial/Documents/fapar.tif</file>
<formatName>GeoTIFF</formatName>
</parameters>
</node>
<applicationData id="Presentation">
<Description/>
<node id="Read">
<displayPosition x="37.0" y="134.0"/>
</node>
<node id="Resample">
<displayPosition x="123.0" y="136.0"/>
</node>
<node id="BiophysicalOp">
<displayPosition x="250.0" y="136.0"/>
</node>
<node id="Write">
<displayPosition x="386.0" y="136.0"/>
</node>
</applicationData>
</graph>
We are doing this computation with the following TILE in input: S2A_MSIL2A_20170214T080021_N0204_R035_T36NXK_20170214T081513.SAFE
I can share the zip, but it’s almost 1gb.
Thanks a lot for the help, please let me know if I should share additional info.