Thanks it works!!
I m using Sentinel-2 L2A product, and I need SCL, WVP and AOT maps.
Using PCI I can see and export easily theses maps (see Fig.1).
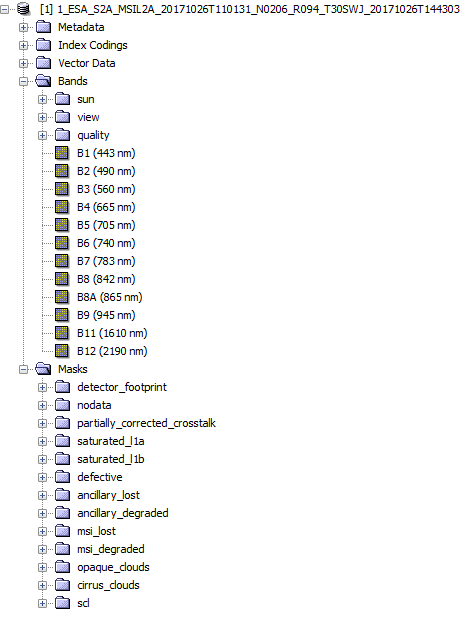
But, with SNAP, I can not find them (Fig.2)
Even when read the available bands using snappy, these bands are absent.
############################################################################
file_name=’/home/ndjamai/job/S2A_L2A/S2A_USER_PRD_MSIL2A_PDMC_20160902T183652_R098_V20160901T173902_20160901T174728.SAFE//S2A_USER_MTD_SAFL2A_PDMC_20160902T183652_R098_V20160901T173902_20160901T174728.xml’
print(“Reading…”)
product = ProductIO.readProduct(file_name)
width = product.getSceneRasterWidth()
height = product.getSceneRasterHeight()
name = product.getName()
description = product.getDescription()
band_names = product.getBandNames()
print(“Product: %s, %s” % (name, description))
print(“Raster size: %d x %d pixels” % (width, height))
print("Start time: " + str(product.getStartTime()))
print("End time: " + str(product.getEndTime()))
print(“Bands: %s” % (list(band_names)))
Product: S2A_USER_MTD_SAFL2A_PDMC_20160902T183652_R098_V20160901T173902_20160901T174728, None
Raster size: 10980 x 10980 pixels
Start time: 01-SEP-2016 17:39:02.026000
End time: 01-SEP-2016 17:47:28.547000
Bands: [‘B2’, ‘B3’, ‘B4’, ‘B5’, ‘B6’, ‘B7’, ‘B8’, ‘B8A’, ‘B11’, ‘B12’, ‘quality_aot’, ‘quality_wvp’, ‘quality_cloud_confidence’, ‘quality_snow_confidence’, ‘quality_scene_classification’, ‘view_zenith_mean’, ‘view_azimuth_mean’, ‘sun_zenith’, ‘sun_azimuth’, ‘view_zenith_B1’, ‘view_azimuth_B1’, ‘view_zenith_B2’, ‘view_azimuth_B2’, ‘view_zenith_B3’, ‘view_azimuth_B3’, ‘view_zenith_B4’, ‘view_azimuth_B4’, ‘view_zenith_B5’, ‘view_azimuth_B5’, ‘view_zenith_B6’, ‘view_azimuth_B6’, ‘view_zenith_B7’, ‘view_azimuth_B7’, ‘view_zenith_B8’, ‘view_azimuth_B8’, ‘view_zenith_B8A’, ‘view_azimuth_B8A’, ‘view_zenith_B9’, ‘view_azimuth_B9’, ‘view_zenith_B10’, ‘view_azimuth_B10’, ‘view_zenith_B11’, ‘view_azimuth_B11’, ‘view_zenith_B12’, ‘view_azimuth_B12’]
######################################################################
I dont know what is the problem,
Fig.1
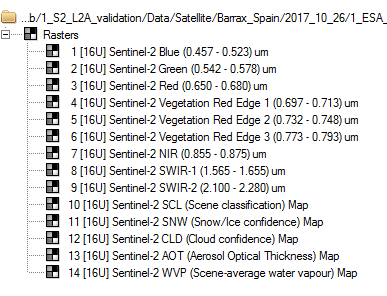
Fig.2