Dear marpet,
when I try SNAP 7.0 to batch sentinel2 with processdata.bash on Mac . but it works well in linux with SNAP8.0 ago. So the processdata only work for SNAP8? Could you give a hand? thanks
By the way, may I change “NetCDF4-CF” to “NetCDF4-BEAM” in processData.bash? thanks
Best
Binbin
error:
Binbin@mMISCLab bin % ./processDataset.bash msi_res20_c2rcc_bin.xml resample_20m.properties /Users/Binbin/Documents/trytry /Users/Binbin/Documents/trtry/
./processDataset.bash: line 51: realpath: command not found
sed: illegal option – r
usage: sed script [-Ealn] [-i extension] [file …]
sed [-Ealn] [-i extension] [-e script] … [-f script_file] … [file …]
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.activator.GDALDistributionInstaller: No distribution folder found on Macintosh.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
Executing processing graph
done.
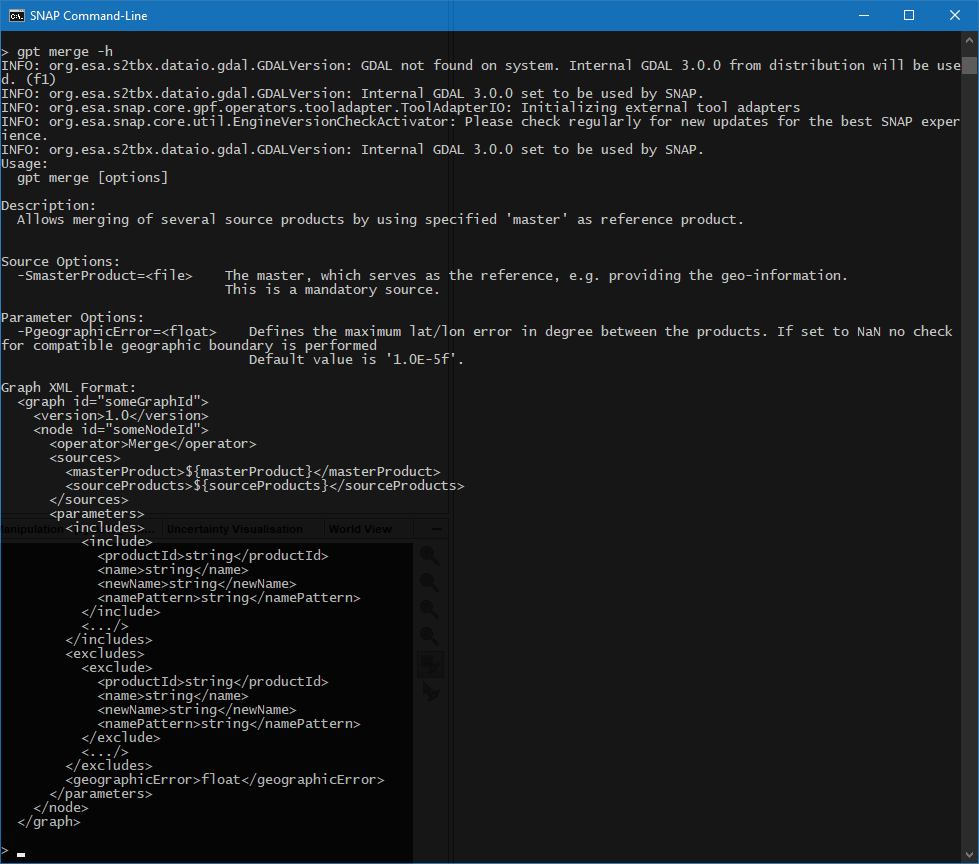
org.esa.snap.core.gpf.graph.GraphException: Missing source ‘${sourceProduct}’ in node ‘S2Resampling’
at org.esa.snap.core.gpf.graph.GraphContext.initNodeDependencies(GraphContext.java:113)
at org.esa.snap.core.gpf.graph.GraphContext.(GraphContext.java:90)
at org.esa.snap.core.gpf.graph.GraphContext.(GraphContext.java:64)
at org.esa.snap.core.gpf.graph.GraphProcessor.executeGraph(GraphProcessor.java:128)
at org.esa.snap.core.gpf.main.DefaultCommandLineContext.executeGraph(DefaultCommandLineContext.java:86)
at org.esa.snap.core.gpf.main.CommandLineTool.executeGraph(CommandLineTool.java:534)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraph(CommandLineTool.java:388)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraphOrOperator(CommandLineTool.java:287)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:188)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:121)
at org.esa.snap.core.gpf.main.GPT.run(GPT.java:54)
at org.esa.snap.core.gpf.main.GPT.main(GPT.java:34)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.esa.snap.runtime.Launcher.lambda$run$0(Launcher.java:55)
at org.esa.snap.runtime.Engine.runClientCode(Engine.java:189)
at org.esa.snap.runtime.Launcher.run(Launcher.java:51)
at org.esa.snap.runtime.Launcher.main(Launcher.java:31)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at com.exe4j.runtime.LauncherEngine.launch(LauncherEngine.java:65)
at com.install4j.runtime.launcher.UnixLauncher.main(UnixLauncher.java:57)
Error: Missing source ‘${sourceProduct}’ in node 'S2Resampling’msi_res20_c2rcc_bin.xml (1.1 KB) processDataset.bash (1.5 KB)resample_20m.properties (55 Bytes)