Hi there,
I’m a new user of Sentinel-2 data and have downloaded a Level-2A product which is cloud free. I’d like to estimate chlorophyll-a (Chl-a) using SNAP, but I’m unsure of the best approach. I know C2RCC is commonly used for this, but from what I’ve read, it requires an L1C product or resampled data. Since my image contains 12 bands with different pixel resolutions, how can I calculate Chl-a from this dataset? Or does SNAP only provide indices for Chl-a estimation?
Additionally, while exploring masking options, I noticed a water mask is available, but I couldn’t find a land mask in this product. Is there a way to apply land masking in SNAP for Sentinel-2 L2A?
Any guidance would be greatly appreciated!
@abruescas @Marco_EOM
Best regards,
Rony
Hi Rony,
Yes, it’s true. C2RCC requires resampled L1C data. You can use the Resampling tool in SNAP. Resample the data set to either 60, 20 or 10 meters. After this preprocessing step you can use the C2RCC. And you should use the original L1C data products in SAFE format as starting point.
A simple tutorial for C2RCC is missing. But in short you use the resampling with the default options. Just choose you resolution. Afterwards you use the C2RCC. You should select the right neural net for your area.
| Name |
Desccription |
| C2RCC-Nets |
Standard NNs preferably to be used for eutrophic to mesotrophic water types. |
| C2X-Nets |
Special NNs for high concentrations of suspended material and chlorophyll concentration. |
| C2X-COMPLEX-Nets |
Special NNs for optically complex water types, preferably to be used for inland waters. |
More details you find on the c2rcc.org website.
The other parameters, like salinity, water temperature, elevation, etc., can be set to further enhance the results. But the impact is in general not as high as the neural net parameter.
The RUS Webinar: Freshwater Quality Monitoring with Sentinel-2 - HYDR02 is a quite complex example for a SNAP beginner. But it shows what is possible with SNAP.
Yes, there is no default land mask. But you can make your own easily. For example, by using the inverted water mask and consider the cloud masks.
!scl_water && !scl_cloud_medium_proba && !scl_cloud_high_proba && !scl_thin_cirrus
See the Mask Manager
You can also use IdePix to generate a separate pixel classification.
IdePix Introduction
1 Like
Thank you for your kind response.
If I understand correctly, LEVEL-2A images cannot be used to estimate Chl-a, and I need to download the L1C images instead.
Could you kindly provide any tutorials or resources on processing Sentinel-2 data in Python? I would greatly appreciate your guidance.
Yes, you need to download your data as L1C images again.
I have blog where I listed 15 great tutorials. Especially. Number 3 and 15 can be of interest for you. The configuration of snappy has slightly changed recently. You need to consider this wiki page: Configure Python to use the new SNAP-Python (esa_snappy) interface (SNAP version 10+)
And if you want to use SNAP with Python you should know its Java API and understand the general concepts Developer Guide - SNAP - Confluence
Thanks for providing this one.
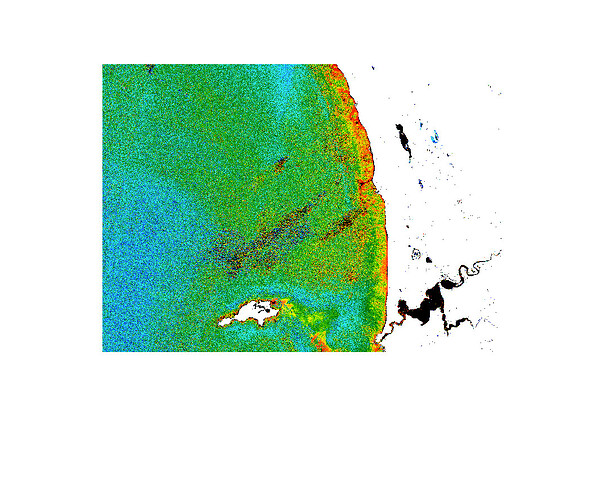
I applied resampling and then processed the L1C image using C2RCC to derive chlorophyll-a concentration. However, I’m unsure about the output—it appears scattered, with pixels looking dispersed rather than forming the smooth contours typically seen in L3 images at 4 km or 1 km resolution.
Could you take a look at the images and help me understand how to interpret them?
Thanks,
Great that you got the results.
Regarding the noise, CHL results for S2 MSI is more noisy than S2 OLCI this is known.
Reasons might be that S2 has been desgined with land application in mind and OLCI was desgined for land and ocean. The signal to noise ratio is probably not so good for water areas. The higher resolution might be also a cause. The noise is smoothed out when creating L3 images. You can smooth your generated image by using the filters provided in SNAP without creaing L3 images. This should result in nicer looking images.
But maybe I’m talking rubbish. Take my comments with a grain of salt. Even I have learned a few things the last years, I’m still a software developer in the first place and not an EO scientist.
Maybe @abruescas can comment and correct me or add additional information.
C2RCC can produce noisy outputs, as you have experienced. S2-MSI has also a bad alignment on the cameras that produces visible stripes over water surfaces sometimes. If you are not going to quantitatively use the outputs, you could apply some filter to smooth the resulting images, so you will have a nicer picture (Raster/Filtered Band). But if you want to use the values, be careful with the smoothing.
2 Likes