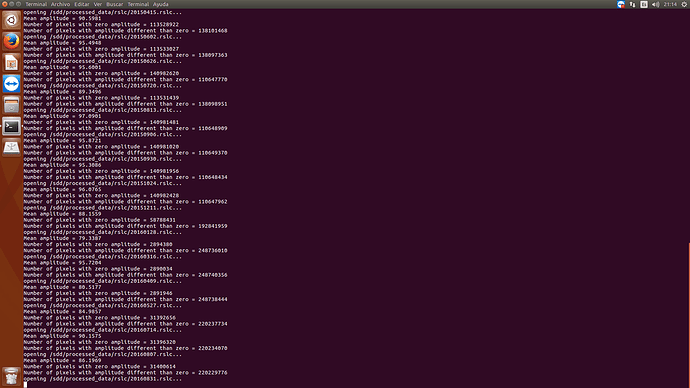
try to rename the folder “processed data” to “processed_data” and run again. It could be that the script expects the variables in the exact number and order and the whitespace caused the error.
good job!
Getting into StaMPS and its preprocessing takes its time, but the possibilities and results are quite rewarding.
I need new results!!! 


Another way to avoid this problem is to pass the path-string in quotation marks, or use \ before the white space to escape it, this can also be used in order to escape signs like $ which do have other meanings in shell scripts…anyway avoiding white space and special sings in file and directory names is always recommend.
HI
To calculation PSI in SNAP 6.0 I did below Steps:
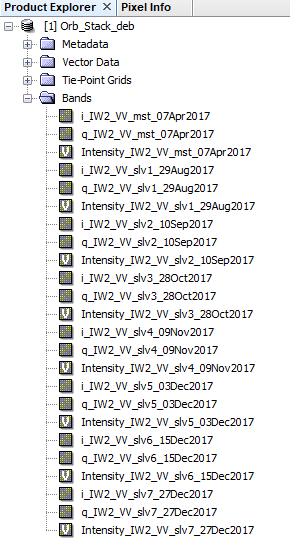
1- Prepared the Orb_Stack_deb
including:
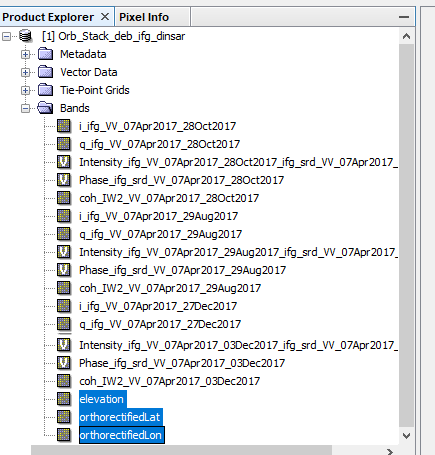
2- prepared the Orb_Stack_deb _inf_dinsar
Including : elevation and orthorectifiedLat and orthorectifiedLon
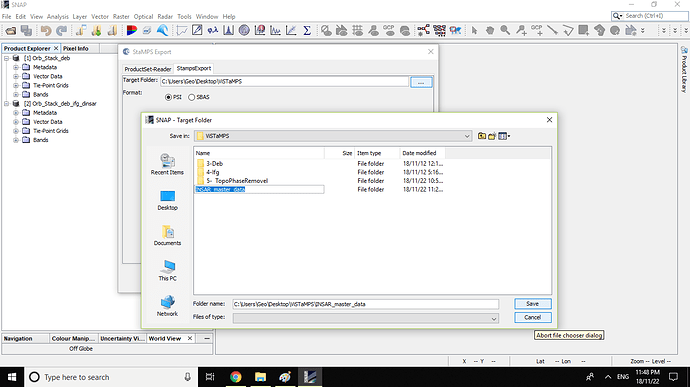
3- Export them by SNAP ‘‘StaMPS export tool’’ into “INSAR_master_data” folder
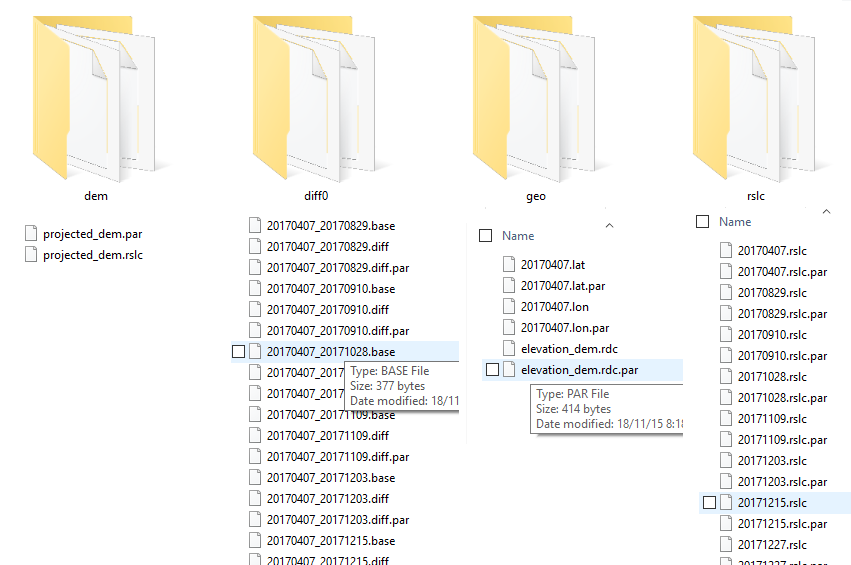
4- Now i have this folders and their contents
5- Please guide me to the next steps of STaMPS
My questions are as follows:
Command mt_prep_gamma (in StaMPS-4.1-beta\bin) In what software and how it runs?
Should I get this folder (13 gigabytes) into linux and run it there? How?
Should I run with Cygwin64 ? How?
The next step in the software should run? How?
Please provide detailed guidance
I read all post in topic but can not STaMPS !!!
Thanks in advance
I don’t recommend cygwin. If you have the chance to use Linux for the next steps, download the StaMPS scripts there and set the variables of the other software (you need matlab, triangle and snaphu, the rest is only used for pre-processing which was done in SNAP)
Once you have these programs installed, you source the config file as described in the StaMPS manual or here:
- Linux Installation using StaMPS and S-1 data
- About the STaMPS category
- Workflow between SNAP and StaMPS
- Problem with StaMPS installation using Linux
mdelgado also explained it well here: How to prepare Sentinel-1 images stack for PSI/SBAS in SNAP 5
If everything is installed, you call the scripts to prepare the data for matlab processing as described in the manual under chapter 6 PS processing.
Hi ABraun and thank U for response
unfortunately, i not know how run " mt_prep_gamma" and which software is need to ran it?
is it possible to you mention the steps after stages above(step by step).
Should I copy these folder (about 13 gigabytes) into linux ?
i copied these folder to linux but can not run " mt_prep_gamma"
thanks
No software, these are linux scripts. You open your shell and type in the commands.
Maybe someone at yout place who is experienced with Linux can assist you. I think this is beyond the scope of this forum, but it has been explained here several times.
Maybe also this is a start.
Hi all @ABraun @thho @Fikretjfm
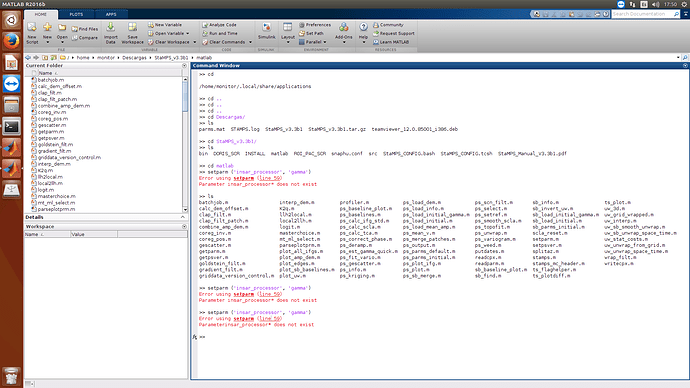
In the end of my processing I have problem with MATLAB, please help! 


Parameter insar_processor* does not exist!!!
Thanks for advance
delete the whitespace before the brackets:
setparm('insar_processor','gamma');
Matlab (and other scripting languages) are quite merciless regarding such small things.
Progressing @ABraun, thanks very much!, was a problem of whitespace…
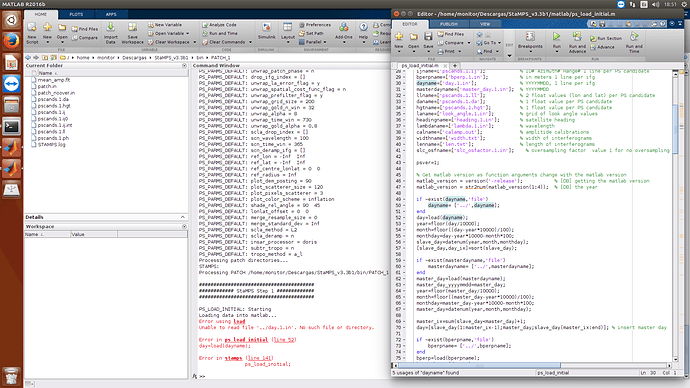
Now, I have new error, in the script ps_load_initial
Hi @gabrielaquintana77,
Please use the latest StaMPS developer version and not latest releasse from https://github.com/dbekaert/stamps , so in such way you may use already the mt_prep_snap and the latest StaMPS version which guarantees the full compatibility between SNAP output with StaMPS PSI processing.
Prior the latest StaMPS version, when I was using previous ones I have also encountered the same issue than you, and at that time I have created the day_1.in and the master_day.in files if they were not present. Sometimes even the headidng.1.in file as well
In addition, using the snap2stamps scripts from https://github.com/mdelgadoblasco/snap2stamps, makes all the DInSAR processing simpler.
Enjoy your processing!
the page seems to have an error at the moment: https://homepages.see.leeds.ac.uk/~earahoo/stamps/
Do you have another source?
It was already posted! They are now also in Github!
in this case I misunderstood the first sentence because it says
and not latest release from https://github.com/dbekaert/stamps
so I thought this was the wrong one.
ouh! yeah, it could be a bit misleading…
There was a new release, but later, on the developer version that you can download from the github as well, they fixed the ps_load_initial_gamma for the cases in which SNAP saved zero values …
This is needed to avoid further issues with index out of bounds in following steps.
So I wanted to say that both official release from this summer and developer versions can be downloaded from github, but get the clone from the site itself instead of the release .
I hope this is clearer now.
Sorry for the confusion
Hi, dear all, I find that it’s no error if I run command “stamps”, but if I rerun codes like “stamps(5,8)” after the first “stamps” finished, when I draw the displacement map using the codes like the following,
case {‘u2d-dmo’}
uw=load(phuwname);
scla=load(sclaname);
ph_all=uw.ph_uw;
ph_all=uw.ph_uw - repmat(scla.C_ps_uw,1,size(uw.ph_uw,2)) - scla.ph_scla;
subtract of oscialtor drift in case of envisat
ph_all = ph_all-ph_unw_eni_osci;
ph_all =-ph_all*55.5/4/3.141592653;
units='mm';
deramping ifgs
[ph_all] = ps_deramp(ps,ph_all);
ph_all(:,master_ix)=0;
clear uw scla
ref_ps=ps_setref;
fig_name = ‘u2d-dmo’;
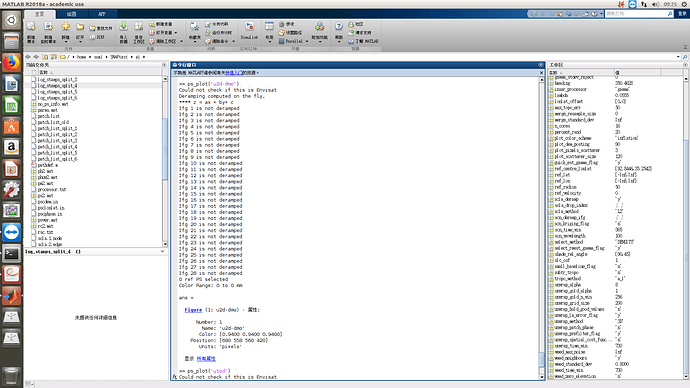
there would be error like the following picture:
Thank you !
according to your screenshots no PS are left.
Please check in the previous steps how many were initially selected and left after the dropping of noisy pixels and weeding.
I have checked the ps points left, about 448000. And I run the command “ps_plot” with the parms using “ps_deramp”, like “ps_plot(‘dmo’)” ,the codes are the following,
case {'u-dmo'}
uw=load(phuwname);
scla=load(sclaname);
ph_all=uw.ph_uw;
ph_all=uw.ph_uw - repmat(scla.C_ps_uw,1,size(uw.ph_uw,2)) - scla.ph_scla;
% subtract of oscialtor drift in case of envisat
ph_all = ph_all-ph_unw_eni_osci;
% deramping ifgs
[ph_all] = ps_deramp(ps,ph_all);
ph_all(:,master_ix)=0;
clear uw scla
ref_ps=ps_setref;
fig_name = 'u-dmo';
and set a breakpoint and see how the values change. I found “scla.C_ps_uw” in line 5 is NAN matrix, as a result, “ph_all” had values before line 5 running, and “ph_all” also become NAN matrix after line 5 computed.