yes, you find instructions in the readme file:
Hi ABraun, I have used the commands on line 88 and 89 of the README file to compile the triangle program. This generated a “showme” and “triangle” file in the triangle folder. A bin folder has not been created. However, I am still running into the same issue in MATLAB as above.
It depends on your operating system how exactly you complile triangle but you need the bin folder and the compiled executable triangle to proceed.
I’m not an exper with the makefiles and compiling, but I hope you find some answers online.
HI ABraun, I have done some testing on the matlab code and it appears that the issue is that the file “snaphu.out” is not being generated during the previous steps. Therefore it is throwing an error when being read in. Do you have any idea what would be causing this / how I could fix it? @mdelgado
Probably snaphu is not installed or not callable from inside MATLAB.
Check that snaphu is correctly installed and put inside the PATH variable reachable from MATLAB.
This should solve it.
Let us know
you could be right. I first thought the error happened in the middle of the unwrapping, but it has obviously not even started. So it could be snaphu indeed.
Apologies, it is my first time compiling programs in C/ playing with linux. I have run the make step on the snaphu makefile and it has created the following folder shown below. I have then added this folder pathway /mainfs/home/hm1r18/snaphu-v2.0.3/bin/ to the StaMPS_CONFIG.bash file:
export TRIANGLE_BIN="/mainfs/home/hm1r18/triangle/bin/"
export SNAPHU_BIN="/mainfs/home/hm1r18/snaphu-v2.0.3/bin/"
However, the same error is still occuring. Could it possible be due to the fact that i am running Matlab 2019a?
No, it cannot be related to the matlab version.
Had you tried to run or call snaphu inside matlab?
Was the StaMPS_CONFIG.bash file sourced before launching/opening Matlab again?
hi Abraun
I have another question! I applied orbit file and backgeoding step, then I used the output files to create stack using coregistration> stack tools> create stack, but I get this error: S1 TOPS Coregistration images must be used
I don’t know what’s the problem:/ Is my way wrong? what should I do then?
@survivere13: There are two ways doing this:
- S1 TOPS Coregistration includes all steps required for the stacking of an image pair for interferometric purposes. However, it is not very memory efficient and usually takes longer than performing the steps subsequently (next option).
- If you want to stack an image pair for interferometric purposes, the correct order is: TOPS Split (select VV only, and reduce the number of bursts whenever possible to the smallest required coverage). Apply Orbit File, then use BackGeocoding to stack the bursts to one product. If you use more than one burst, use Enhanced Spectral Diversity to increase the quality of the coregistration. No further operators are then required.
A revised tutorial for InSAR processing of TOPS data will soon be uploaded.
here it is: http://step.esa.int/docs/tutorials/S1TBX%20TOPSAR%20Interferometry%20with%20Sentinel-1%20Tutorial_v2.pdf
Thanks for the very nice new tutorial!
I may have some comment regarding the suggestions for further analysis, as doing the average displacement you can mitigate the atmospheric effects, but you also assume linear deformation for the entire period, which may not be always the case.
Good job! I like it. Nice to see that the manuals get updated. 
thank you for the feedback! I will include your comment in the tutorial and re-submit it once I have collected some more points.
Hello friends
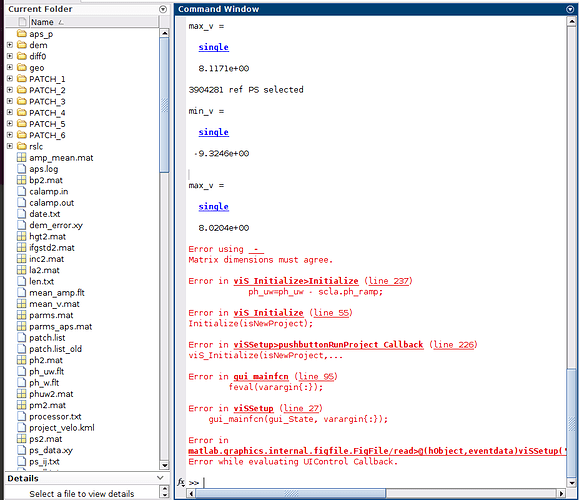
I am working on ubuntu, and I want to visualize results of interferogrmas, deformations, speeds, etc., but when I run viStaMPS in matlab, I get this error message,
Anyone know why the error?
Hello,ABraun,the “INSAR_20150419” folder in the following command needs to be created manually, right?
Yes. And at best executed in a separate folder, not directly inside the INSAR folder to keep the files clean
This error appears for me as well, when creating stack of one burts infgm, however, two burts run without any error and create stacking. Any idea!
If you use Split separately, the next step to stack the products is BackGeocoding, not with the “Create Stack” operator.
No, I’m talking about creating stack of inteferograms,
Apply backgeocoding to create Stack of infgms, gives an error of double rslc of the products, throughout StaMPS export.
sorry, this was not clear to me. I haven’t used the manual preparation a long while so I’m afraid I can’t tell what is the most elegant way to do it at the moment.