use the full_path of data works for me,thanks
Does it strickly require Kubuntu distribution?
I already have Ubuntu with python version 3.8 installed, will that work properly with Stamps?
Hi @Shadi1, Ubuntu is just fine  Kubuntu was just a recommendation for all how come from Windows and like to start off with a more familiar desktop layout
Kubuntu was just a recommendation for all how come from Windows and like to start off with a more familiar desktop layout
hello everyone!
sorry, I am new in PS. Is there anyone can help me to explain about “Unwrap_time_win” and “unwrap_gold_alpha” ?
for the “unwrap_gold_alpha”, I have changed from 0.2, 0.4, … to 1.8, but there’s no change or happen in the result value.
thank you
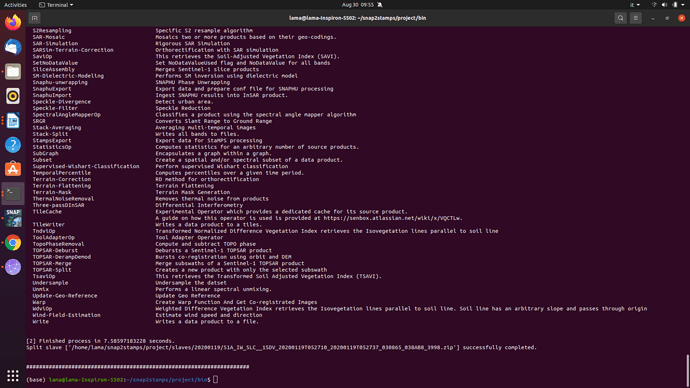
The parameters are explained here: StaMPS/2-4_StaMPS-steps.md · master · Matthias Schlögl / gis-blog · GitLab
Hello, I want to know why initial gamma threshold: 1.000 at d appears_ A=0.05 to 1.000 at D_ A = 0.40, I think the key problem is why it is 1.000, but I don’t know how to solve it.
Because when warning: not enough random phase pixels to set gamma threshold - using default threshold of 0.3 appears, 0 PS selected initially will not appear when the software uses the default index
Dear ABraun,
This worked perfectly for 21 images. But when I upgraded the data set to 50, the debursting took very long time. Hence, I stoped the debursting and created indivduale inteferograms, but I do not know how to put the interferograms in a stack, could you help me please?.
I tried to create a stack from Stack tools but the resulted stack kept the interferograms out of the stack!
Thanks alot.
have you considered using the snap2stamps python scripts? They automatically preprocess the data as required by stamps and because each image is processed separately, there are no memory errors.
Doing it manually is always prone to errors and lack of processing capacities.
I did it manually because I rised the memory capcity to 13G out of 15G, but I had always problems like GC Limitation and Buffer constracting (tha data set is 50 images of Sentinel1 SLC). I tried the scripts, for snap 6 it was OK but for snap8 even the splitting slaves script did not work. Creating each interferogram manually was my only choice.
Best
especially when you are short on RAM you should consider snap2stamps, because every split or coregistration is performed separately (and memory is cleared afterwards) by the scripts.
The splitting should work regardless of the version, which error did you get?
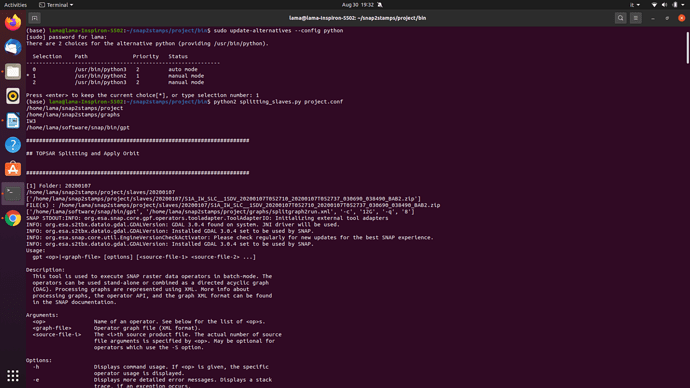
The problem is that the script creat a folder called split with empty sub folders of the slaves names.
Before this I had a problem with the subprocess file:
process = subprocess.Popen(args,stdout=subprocess.PIPE,stderr=subprocess.STDOUT)
Then, I edited this line to:
process = subprocess.Popen(args,stdout=subprocess.PIPE,shell=True,stderr=subprocess.STDOUT).
It worked just to create empty files. I am working on ubunto 20.04.
the fact that it lists all snap commands indicates that the command was not handed over to the shell correctly. Can you please scroll up to the error message?
I could not recognize an error message, this is what makes me stop trying using the scripts. All the commands are listed and a sucessful message appears and the split file only contains empty named folders.
the fact that it lists the commands indicates that the syntax was not correct. Did you first execute slaves_prep.py as a first step? This distributes all zip files in the correct folder structure.
Of coures slaves_prep is working. I have an idea: if I removed the edit of (shell=True) in the line of subprocess.Popen, then I will go back to the error of subprocess file. How could I deal with this error please?.
Just wondering if you use python 2.7 or python 3.X coming by default on Ubuntu 20.04
if the latter, probably the subprocess function has changed in the meanwhile.
Can you please confirm that you are using python 2.7?
The default is python3 but I installed python 2.7 and I switched to it before starting using the scripts.
Dear all
The problem has been solved. The master was splitted out of the master folder. So the master folder was empty. Sorry and thank you so much!.
Great that you found it yourself, good job!