LAI_fAPAR_20to10_GPT_TIFF.xml (3.2 KB)
Dear Support Team,
There is an issue in computing and analysing LAI bands in the processed LAI from L2A S2. In the previous computation, it was easily selected and identified the LAI by changing in the .xml, but now it is not working and it is showing 2 bands in the computed LAI.
Optical Toolbox Kit Module: Version: 10.0.4
Thanks
Dear user,
Could you attach a screenshot of the resulting product opened in SNAP?
Thank you!
Please find the attached screenshot.
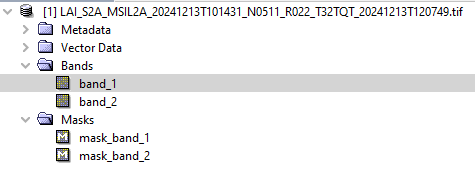
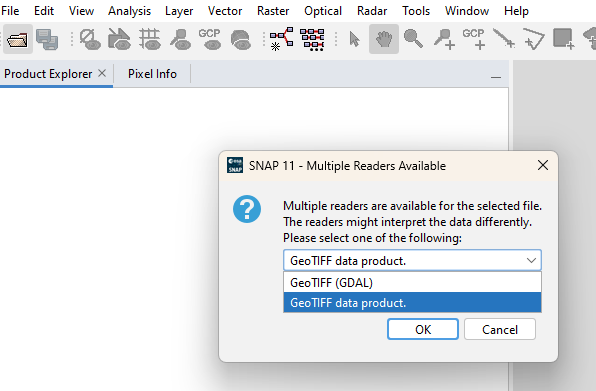
It seems to be an issue with the reader.
When you open the product, SNAP will suggest two readers. Please choose the second one
Thank you for the reply, Actually I am accessing the toolbox by command prompt,
e.g.
for image in /data/SENTINEL-2/L2A/"$year"/*_"$tile_name".zip; do
path="${image:58}"
output_name="LAI_${path}"
output_name="${output_name:0:-4}"
~/esa-snap/bin/gpt /EO_Data/S2/LAI_fAPAR_20to10_GPT_TIFF.xml \
-PinputFile=$image \
-PoutputLAIFile="/EO_Data/S2/$tile_name/BIOPARS_raw/2024/${output_name}.tif" \
-Psensor=S2A
rm -r /home/absingh/.snap/var/cache/s2tbx/l2a-reader/10.0.4/*
done
You can also force GPT to use a specific reader or writer by setting the value of the formatName parameter:
-
for the Read operator:
<node id=“Read”>
<operator>Read</operator>
<sources/>
<parameters class=“com.bc.ceres.binding.dom.XppDomElement”>
<useAdvancedOptions>false</useAdvancedOptions>
<file>C:\Users\zzz\input.tif</file>
<formatName>GeoTIFF</formatName>
<copyMetadata>true</copyMetadata>
<bandNames/>
<pixelRegion>0,0,10980,10980</pixelRegion>
<maskNames/>
</parameters>
</node>
-
for the Write operator:
<node id=“Write”>
<operator>Write</operator>
…
<parameters class=“com.bc.ceres.binding.dom.XppDomElement”>
<file>C:\Users\zzz\output.tif</file>
<formatName>GeoTIFF</formatName>
</parameters>
</node>
Thank for the Reply. Still I am facing the same issue even after setting the format name.
Could you list all the processing steps?
Did you set the format name for the Write operator or for the Read operator?
The format needs to be specified every time you read the product.
Thank you for the reply.
I am using L2A product, and resampled the spectral bands to 10 m before applying the Biophysical LAI processor.
I have also applied the format name in the read and write operator. (attached .xml for your reference)
LAI_fAPAR_20to10_GPT_TIFF-AS.xml (3.4 KB)
Command:
for year in 2024; do
for tile_name in T32TQT; do
for image in /data/SENTINEL-2/L2A/"$year"/*_"$tile_name"_202411*.zip; do
path=$(basename "$image")
output_name="LAI_${path%.zip}"
~/esa-snap/bin/gpt /data/S2/LAI_fAPAR_20to10_GPT_TIFF-AS.xml \
-PinputFile="$image" \
-PoutputLAIFile="/data/S2/$tile_name/BIOPARS_raw/2024/${output_name}.tif" \
-Psensor=S2A
rm -r /home/absingh/.snap/var/cache/s2tbx/l2a-reader/10.0.4/*
done
done
done

