Hello,
i was trying to write a DIMAP product (which can be opened in SNAP without any issues) as a BigTIF-GeoTIFF file via Linux commandline (" /opt/snap9/bin/gpt beam_tiff_conv.xml -q 16 20210214T054409_ifg.tif.dim "), using an XML which i created using the SNAP Graphbuilder:
<graph id="Graph">
<version>1.0</version>
<node id="Read">
<operator>Read</operator>
<sources/>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<useAdvancedOptions>false</useAdvancedOptions>
<file>20210214T054409_ifg.tif.dim</file>
<formatName>BEAM-DIMAP</formatName>
<copyMetadata>true</copyMetadata>
<bandNames/>
<pixelRegion>0,0,16414,18871</pixelRegion>
<maskNames/>
</parameters>
</node>
<node id="Write">
<operator>Write</operator>
<sources>
<sourceProduct refid="Read"/>
</sources>
<parameters class="com.bc.ceres.binding.dom.XppDomElement">
<file>output.tif</file>
<formatName>GeoTIFF-BigTIFF</formatName>
</parameters>
</node>
<applicationData id="Presentation">
<Description/>
<node id="Read">
<displayPosition x="37.0" y="134.0"/>
</node>
<node id="Write">
<displayPosition x="455.0" y="135.0"/>
</node>
</applicationData>
</graph>
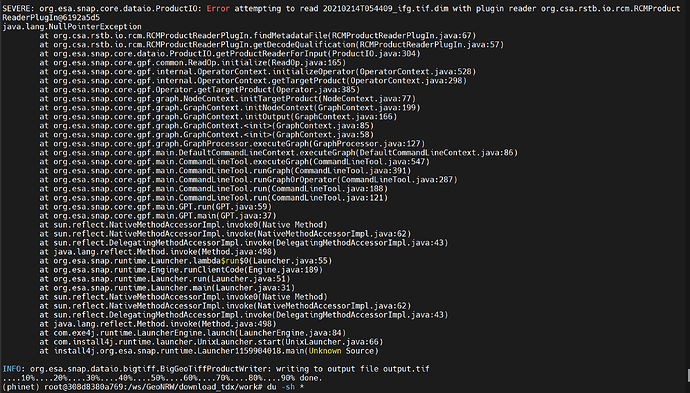
While i was able to execute the command, i got the following respond:
Although, according to the commandline the process was completed, a corrupted .tif file was created, which i was not able to open up in SNAP. The error was the following:
After changing the commandline to: " /opt/snap9/bin/gpt beam_tiff_conv.xml -c 32G -q 16 20210214T054409_ifg.tif.dim "
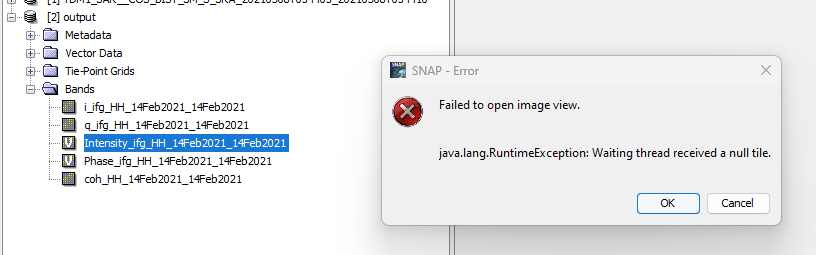
SNAP would show me another error:
I already tried converting the file in the SNAP GUI it self, using the Graphbuilder, however, this also gave me the error shown above.
Is there any way to fix this issue or is this a problem with SNAP?
Thank you!