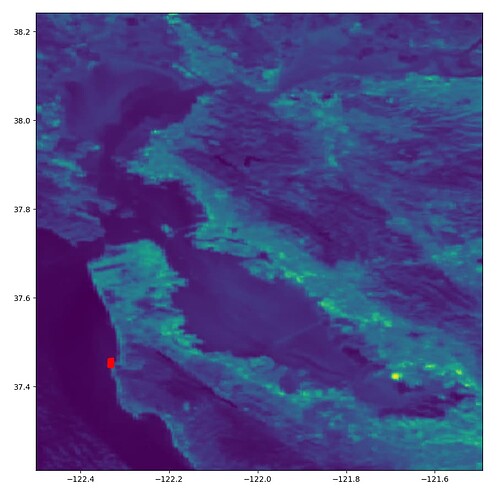
Hello,
I am pretty new to using ESA products. I am downloading S3 OLCI images using the OData API, subsetting my Area of Interest, and applying C2RCC correction using the snappy module in Python. The whole process goes smoothly, but when I load the raster saved as a .tif using rasterio I notice there is a right shift in the longitude and a down shift in the latitude. For example, the red point should be closer to the coordinates (-122.5,37.6) than what it’s suggested by rasterio.
I tried exporting the raster to csv and it is shifted as well in that case. Is there a reprojection that needs to happen somewhere in my processing chain? Here’s my Python code if that helps at all:
import sys
import os
import re
sys.path.append("/home/jarroyoe/.snap/snap-python")
import esa_snappy as snap
sfe_polygon = "POLYGON((-121.92416953468114 37.40082853249865,-121.92416953468114 38.15500781181325,-122.52040601399796 38.15500781181325,-122.52040601399796 37.40082853249865,-121.92416953468114 37.40082853249865))"
sfe_geometry = snap.WKTReader().read(sfe_polygon)
raw_data_path = "/home/jarroyoe/process_bands/SFE/Raw_Data"
processed_data_path = "/home/jarroyoe/process_bands/SFE/Processed_Data"
SFE_rasters = os.listdir(raw_data_path)
for file in SFE_rasters:
raw_raster = snap.ProductIO.readProduct(raw_data_path+"/"+file)
#Process only the SFE
parameters = snap.HashMap()
parameters.put('copyMetadata',True)
parameters.put('geoRegion',sfe_geometry)
sfe_raster = snap.GPF.createProduct('Subset', parameters, raw_raster)
#Perform C2RCC correction
parameters = snap.HashMap()
parameters.put('outputRhown',False)
parameters.put('outputKd',False)
parameters.put('outputUncertainties',False)
c2rcc_corrected_raster = snap.GPF.createProduct('c2rcc.olci', parameters, sfe_raster)
#Extract only relevant bands
parameters = snap.HashMap()
parameters.put('copyMetadata',True)
parameters.put('sourceBands','rtoa_8,rtoa_11,rtoa_18')
relevant_bands = snap.GPF.createProduct('Subset', parameters, c2rcc_corrected_raster)
#Write relevant bands
file_date_position = re.search("[0-9]{8,8}",file).start()
file_date = file[file_date_position:file_date_position+8]
snap.ProductIO.writeProduct(relevant_bands,processed_data_path+"/"+"sfe_bands_8_11_18_"+file_date,'GeoTIFF')
Thanks!